Cross-correlation QC (all)
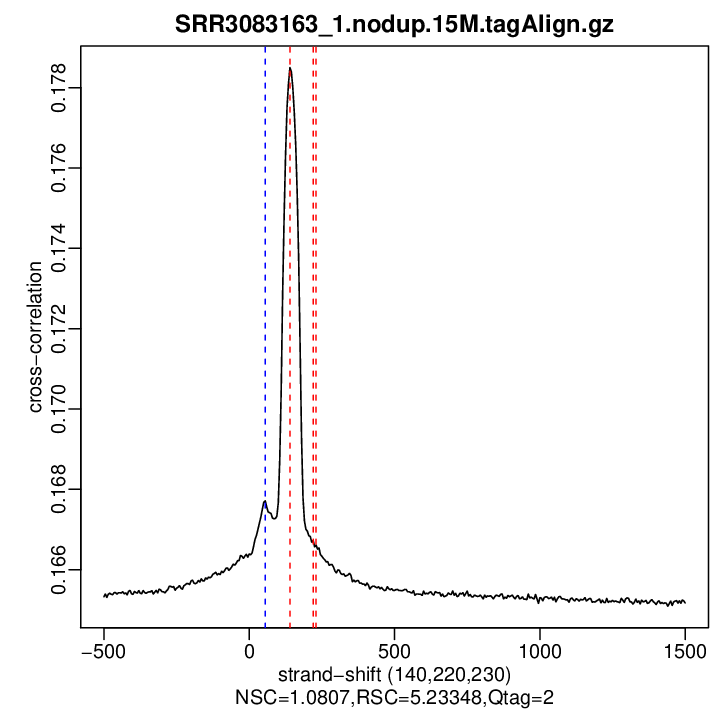
rep1
| numReads | estFragLen | corr_estFragLen | PhantomPeak | corr_phantomPeak | argmin_corr | min_corr | NSC | RSC | |
|---|---|---|---|---|---|---|---|---|---|
| rep1 | 14155684 | 140 | 0.178498977156904 | 55 | 0.1677163 | 1500 | 0.1651692 | 1.080703 | 5.233481 |
Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
Relative strand cross-correlation coefficient (RSC) = col10 in outFile
Estimated fragment length = col3 in outFile, take the top value
Important columns highlighted, but all/whole file can be stored for display

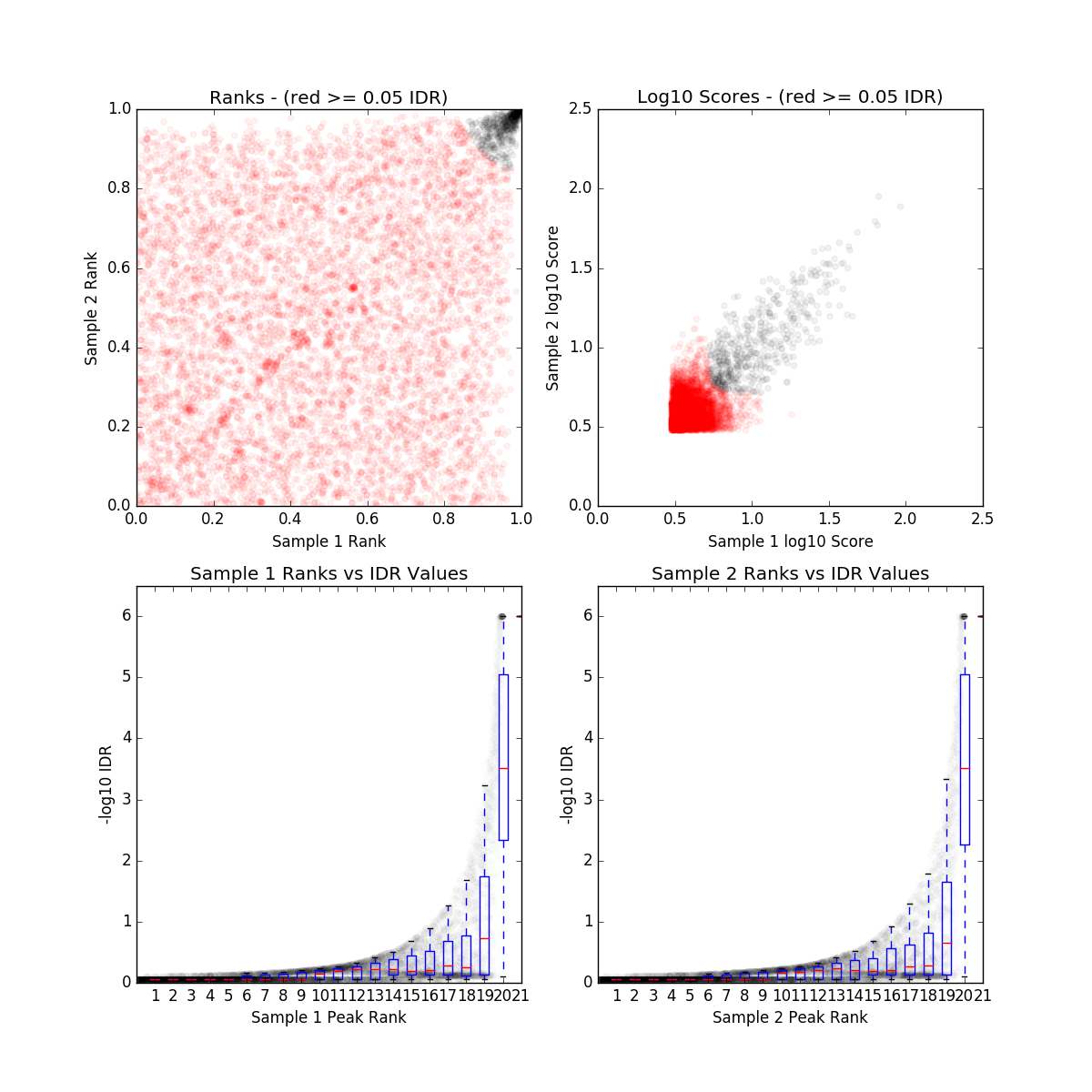
IDR QC (idr)
ZNF30_IDR_final.qc
| Nt | N1 | N2 | Np | conservative_set | optimal_set | rescue_ratio | self_consistency_ratio | reproducibility |
|---|---|---|---|---|---|---|---|---|
| 0 | 482 | 0 | 0 | N/A | N/A | NaN | 1.0 | 1 |