Cross-correlation QC (all)
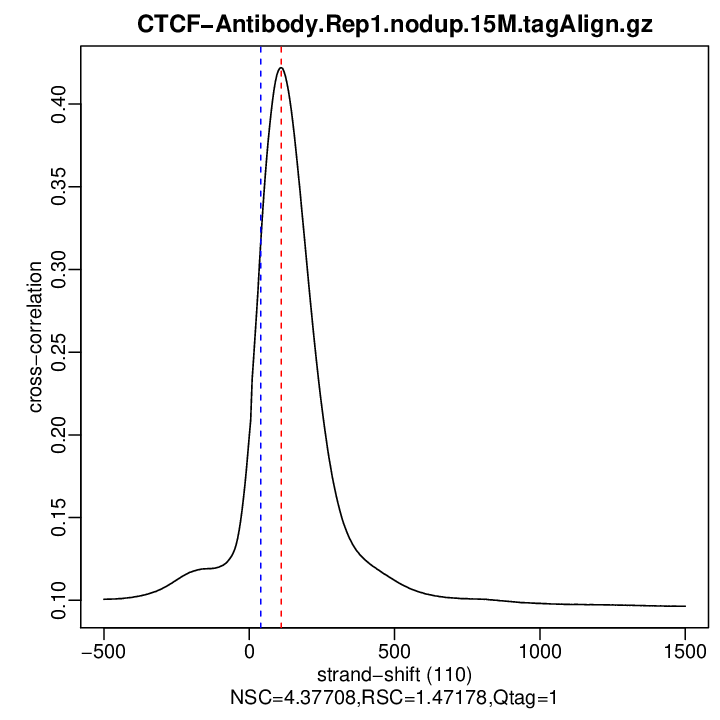
rep1 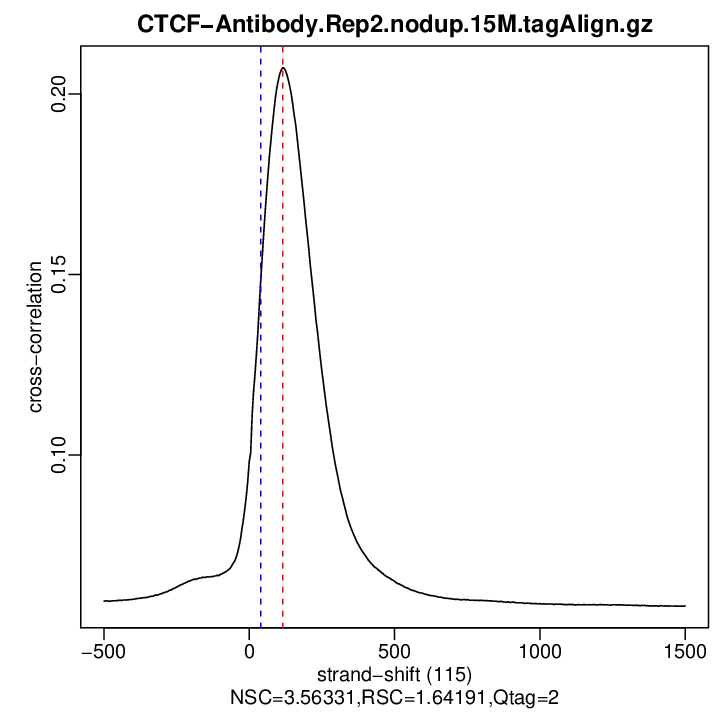
rep2
| numReads | estFragLen | corr_estFragLen | PhantomPeak | corr_phantomPeak | argmin_corr | min_corr | NSC | RSC | |
|---|---|---|---|---|---|---|---|---|---|
| rep1 | 10397826 | 110 | 0.422063186386812 | 40 | 0.3176792 | 1500 | 0.09642573 | 4.377081 | 1.471785 |
| rep2 | 4803904 | 115 | 0.207286177158335 | 40 | 0.1489896 | 1500 | 0.05817234 | 3.563312 | 1.64191 |
Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
Relative strand cross-correlation coefficient (RSC) = col10 in outFile
Estimated fragment length = col3 in outFile, take the top value
Important columns highlighted, but all/whole file can be stored for display

