| p-value: | 1e-100 |
| log p-value: | -2.307e+02 |
| Information Content per bp: | 1.877 |
| Number of Target Sequences with motif | 1707.0 |
| Percentage of Target Sequences with motif | 40.81% |
| Number of Background Sequences with motif | 5314.9 |
| Percentage of Background Sequences with motif | 25.69% |
| Average Position of motif in Targets | 394.3 +/- 348.9bp |
| Average Position of motif in Background | 285.8 +/- 206.7bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.53 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
hb/dmmpmm(SeSiMCMC)/fly
| Match Rank: | 1 |
| Score: | 0.91
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | RAAAAAAA
-AAAAAA- |
|


|
|
PB0192.1_Tcfap2e_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.90
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----RAAAAAAA-
TACTGGAAAAAAAA |
|


|
|
hb/dmmpmm(Down)/fly
| Match Rank: | 3 |
| Score: | 0.89
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -RAAAAAAA
CATAAAAAA |
|


|
|
PB0182.1_Srf_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.88
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----RAAAAAAA-----
GTTAAAAAAAAAAATTA |
|


|
|
hb/MA0049.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.87
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --RAAAAAAA
GCATAAAAAA |
|


|
|
hb/dmmpmm(Pollard)/fly
| Match Rank: | 6 |
| Score: | 0.87
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | RAAAAAAA
ATAAAAAA |
|


|
|
hb/dmmpmm(Noyes)/fly
| Match Rank: | 7 |
| Score: | 0.87
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --RAAAAAAA--
ACNAAAAAAACA |
|


|
|
hb/dmmpmm(Bigfoot)/fly
| Match Rank: | 8 |
| Score: | 0.84
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -RAAAAAAA
CATAAAAAA |
|


|
|
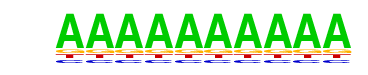
SeqBias: polyA-repeat
| Match Rank: | 9 |
| Score: | 0.84
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | RAAAAAAA--
AAAAAAAAAA |
|

|
|
hb/dmmpmm(Papatsenko)/fly
| Match Rank: | 10 |
| Score: | 0.83
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -RAAAAAAA
CAAAAAAA- |
|


|
|





















