| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | ABF1/SacCer-Promoters/Homer | 1e-180 | -4.156e+02 | 0.0000 | 1131.0 | 34.03% | 3056.4 | 14.21% | motif file (matrix) |
pdf |
| 2 |  | REB1/SacCer-Promoters/Homer | 1e-152 | -3.520e+02 | 0.0000 | 696.0 | 20.94% | 1469.4 | 6.83% | motif file (matrix) |
pdf |
| 3 |  | SeqBias: CG-repeat | 1e-54 | -1.249e+02 | 0.0000 | 669.0 | 20.13% | 2335.7 | 10.86% | motif file (matrix) |
pdf |
| 4 |  | RLR1?/SacCer-Promoters/Homer | 1e-51 | -1.197e+02 | 0.0000 | 1366.0 | 41.10% | 6175.9 | 28.71% | motif file (matrix) |
pdf |
| 5 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-46 | -1.060e+02 | 0.0000 | 458.0 | 13.78% | 1449.3 | 6.74% | motif file (matrix) |
pdf |
| 6 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-37 | -8.707e+01 | 0.0000 | 528.0 | 15.88% | 1907.1 | 8.87% | motif file (matrix) |
pdf |
| 7 |  | bHLHE40(bHLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-35 | -8.250e+01 | 0.0000 | 339.0 | 10.20% | 1043.0 | 4.85% | motif file (matrix) |
pdf |
| 8 |  | Cbf1(bHLH)/Yeast-Cbf1-ChIP-Seq(GSE29506)/Homer | 1e-34 | -7.966e+01 | 0.0000 | 313.0 | 9.42% | 943.4 | 4.39% | motif file (matrix) |
pdf |
| 9 |  | DPL-1(E2F)/cElegans-Adult-ChIP-Seq(modEncode)/Homer | 1e-34 | -7.830e+01 | 0.0000 | 824.0 | 24.79% | 3538.3 | 16.45% | motif file (matrix) |
pdf |
| 10 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-32 | -7.468e+01 | 0.0000 | 435.0 | 13.09% | 1542.2 | 7.17% | motif file (matrix) |
pdf |
| 11 |  | SFP1/SacCer-Promoters/Homer | 1e-32 | -7.462e+01 | 0.0000 | 438.0 | 13.18% | 1557.5 | 7.24% | motif file (matrix) |
pdf |
| 12 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-30 | -6.961e+01 | 0.0000 | 302.0 | 9.09% | 950.8 | 4.42% | motif file (matrix) |
pdf |
| 13 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-29 | -6.800e+01 | 0.0000 | 367.0 | 11.04% | 1262.6 | 5.87% | motif file (matrix) |
pdf |
| 14 |  | Max(bHLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-27 | -6.283e+01 | 0.0000 | 457.0 | 13.75% | 1744.3 | 8.11% | motif file (matrix) |
pdf |
| 15 |  | PIF5ox(bHLH)/Arabidopsis-PIF5ox-ChIP-Seq(GSE35062)/Homer | 1e-24 | -5.620e+01 | 0.0000 | 673.0 | 20.25% | 2954.4 | 13.73% | motif file (matrix) |
pdf |
| 16 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-24 | -5.547e+01 | 0.0000 | 627.0 | 18.86% | 2711.3 | 12.60% | motif file (matrix) |
pdf |
| 17 |  | PIF4(bHLH)/Seedling-PIF4-ChIP-Seq(GSE35315)/Homer | 1e-23 | -5.415e+01 | 0.0000 | 739.0 | 22.23% | 3345.1 | 15.55% | motif file (matrix) |
pdf |
| 18 |  | E-box/Arabidopsis-Promoters/Homer | 1e-22 | -5.166e+01 | 0.0000 | 408.0 | 12.27% | 1593.4 | 7.41% | motif file (matrix) |
pdf |
| 19 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-22 | -5.117e+01 | 0.0000 | 1044.0 | 31.41% | 5145.5 | 23.92% | motif file (matrix) |
pdf |
| 20 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-21 | -4.957e+01 | 0.0000 | 338.0 | 10.17% | 1259.1 | 5.85% | motif file (matrix) |
pdf |
| 21 |  | FHY3(FAR1)/Arabidopsis-FHY3-ChIP-Seq(GSE30711)/Homer | 1e-20 | -4.612e+01 | 0.0000 | 329.0 | 9.90% | 1243.5 | 5.78% | motif file (matrix) |
pdf |
| 22 |  | Egr2(Zf)/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-18 | -4.187e+01 | 0.0000 | 110.0 | 3.31% | 270.6 | 1.26% | motif file (matrix) |
pdf |
| 23 |  | SPCH(bHLH)/Seedling-SPCH-ChIP-Seq(GSE57497)/Homer | 1e-18 | -4.163e+01 | 0.0000 | 824.0 | 24.79% | 4006.5 | 18.63% | motif file (matrix) |
pdf |
| 24 |  | NPAS2(bHLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-17 | -3.955e+01 | 0.0000 | 606.0 | 18.23% | 2793.7 | 12.99% | motif file (matrix) |
pdf |
| 25 |  | E-box(bHLH)/Promoter/Homer | 1e-16 | -3.770e+01 | 0.0000 | 96.0 | 2.89% | 232.7 | 1.08% | motif file (matrix) |
pdf |
| 26 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-14 | -3.386e+01 | 0.0000 | 281.0 | 8.45% | 1111.5 | 5.17% | motif file (matrix) |
pdf |
| 27 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-14 | -3.230e+01 | 0.0000 | 1058.0 | 31.83% | 5564.6 | 25.87% | motif file (matrix) |
pdf |
| 28 |  | TATA-box/SacCer-Promoters/Homer | 1e-14 | -3.227e+01 | 0.0000 | 1257.0 | 37.82% | 6783.1 | 31.53% | motif file (matrix) |
pdf |
| 29 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo_et_al.)/Homer | 1e-13 | -3.151e+01 | 0.0000 | 793.0 | 23.86% | 3996.6 | 18.58% | motif file (matrix) |
pdf |
| 30 |  | SUT1?/SacCer-Promoters/Homer | 1e-11 | -2.685e+01 | 0.0000 | 2306.0 | 69.37% | 13692.1 | 63.65% | motif file (matrix) |
pdf |
| 31 |  | TOD6?/SacCer-Promoters/Homer | 1e-10 | -2.491e+01 | 0.0000 | 227.0 | 6.83% | 923.0 | 4.29% | motif file (matrix) |
pdf |
| 32 |  | ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer | 1e-10 | -2.482e+01 | 0.0000 | 239.0 | 7.19% | 986.6 | 4.59% | motif file (matrix) |
pdf |
| 33 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-10 | -2.465e+01 | 0.0000 | 129.0 | 3.88% | 441.1 | 2.05% | motif file (matrix) |
pdf |
| 34 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-10 | -2.313e+01 | 0.0000 | 569.0 | 17.12% | 2843.1 | 13.22% | motif file (matrix) |
pdf |
| 35 |  | KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer | 1e-9 | -2.225e+01 | 0.0000 | 422.0 | 12.70% | 2017.2 | 9.38% | motif file (matrix) |
pdf |
| 36 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-8 | -1.995e+01 | 0.0000 | 239.0 | 7.19% | 1043.0 | 4.85% | motif file (matrix) |
pdf |
| 37 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-8 | -1.993e+01 | 0.0000 | 435.0 | 13.09% | 2130.2 | 9.90% | motif file (matrix) |
pdf |
| 38 |  | ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer | 1e-8 | -1.878e+01 | 0.0000 | 46.0 | 1.38% | 112.7 | 0.52% | motif file (matrix) |
pdf |
| 39 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-8 | -1.864e+01 | 0.0000 | 354.0 | 10.65% | 1693.0 | 7.87% | motif file (matrix) |
pdf |
| 40 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-8 | -1.855e+01 | 0.0000 | 214.0 | 6.44% | 926.4 | 4.31% | motif file (matrix) |
pdf |
| 41 |  | KLF10(Zf)/HEK293-KLF10.GFP-ChIP-Seq(GSE58341)/Homer | 1e-8 | -1.849e+01 | 0.0000 | 185.0 | 5.57% | 774.7 | 3.60% | motif file (matrix) |
pdf |
| 42 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-7 | -1.796e+01 | 0.0000 | 124.0 | 3.73% | 469.7 | 2.18% | motif file (matrix) |
pdf |
| 43 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-7 | -1.689e+01 | 0.0000 | 150.0 | 4.51% | 610.6 | 2.84% | motif file (matrix) |
pdf |
| 44 |  | GAGA-repeat/Arabidopsis-Promoters/Homer | 1e-6 | -1.480e+01 | 0.0000 | 378.0 | 11.37% | 1899.4 | 8.83% | motif file (matrix) |
pdf |
| 45 |  | Klf9(Zf)/GBM-Klf9-ChIP-Seq(GSE62211)/Homer | 1e-6 | -1.449e+01 | 0.0000 | 98.0 | 2.95% | 371.8 | 1.73% | motif file (matrix) |
pdf |
| 46 |  | SeqBias: TA-repeat | 1e-6 | -1.396e+01 | 0.0000 | 2746.0 | 82.61% | 17058.3 | 79.30% | motif file (matrix) |
pdf |
| 47 |  | HIF-1a(bHLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-5 | -1.350e+01 | 0.0000 | 348.0 | 10.47% | 1752.7 | 8.15% | motif file (matrix) |
pdf |
| 48 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-5 | -1.275e+01 | 0.0000 | 1198.0 | 36.04% | 6951.8 | 32.32% | motif file (matrix) |
pdf |
| 49 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-5 | -1.211e+01 | 0.0000 | 522.0 | 15.70% | 2808.0 | 13.05% | motif file (matrix) |
pdf |
| 50 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-5 | -1.210e+01 | 0.0000 | 82.0 | 2.47% | 313.1 | 1.46% | motif file (matrix) |
pdf |
| 51 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.196e+01 | 0.0000 | 753.0 | 22.65% | 4212.2 | 19.58% | motif file (matrix) |
pdf |
| 52 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-5 | -1.180e+01 | 0.0001 | 451.0 | 13.57% | 2393.9 | 11.13% | motif file (matrix) |
pdf |
| 53 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-5 | -1.170e+01 | 0.0001 | 95.0 | 2.86% | 383.0 | 1.78% | motif file (matrix) |
pdf |
| 54 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-4 | -1.138e+01 | 0.0001 | 43.0 | 1.29% | 134.6 | 0.63% | motif file (matrix) |
pdf |
| 55 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-4 | -1.101e+01 | 0.0001 | 364.0 | 10.95% | 1899.9 | 8.83% | motif file (matrix) |
pdf |
| 56 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-4 | -9.723e+00 | 0.0004 | 147.0 | 4.42% | 683.5 | 3.18% | motif file (matrix) |
pdf |
| 57 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer | 1e-4 | -9.580e+00 | 0.0005 | 41.0 | 1.23% | 136.4 | 0.63% | motif file (matrix) |
pdf |
| 58 |  | HIF2a(bHLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer | 1e-4 | -9.446e+00 | 0.0005 | 518.0 | 15.58% | 2860.1 | 13.30% | motif file (matrix) |
pdf |
| 59 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-4 | -9.354e+00 | 0.0006 | 393.0 | 11.82% | 2112.5 | 9.82% | motif file (matrix) |
pdf |
| 60 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-4 | -9.345e+00 | 0.0006 | 985.0 | 29.63% | 5745.9 | 26.71% | motif file (matrix) |
pdf |
| 61 |  | NRF(NRF)/Promoter/Homer | 1e-4 | -9.270e+00 | 0.0006 | 78.0 | 2.35% | 320.0 | 1.49% | motif file (matrix) |
pdf |
| 62 |  | Sp1(Zf)/Promoter/Homer | 1e-3 | -9.028e+00 | 0.0007 | 43.0 | 1.29% | 149.1 | 0.69% | motif file (matrix) |
pdf |
| 63 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.754e+00 | 0.0010 | 611.0 | 18.38% | 3449.4 | 16.04% | motif file (matrix) |
pdf |
| 64 |  | PAX5(Paired,Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-3 | -8.391e+00 | 0.0014 | 163.0 | 4.90% | 794.7 | 3.69% | motif file (matrix) |
pdf |
| 65 |  | E2F(E2F)/Hela-CellCycle-Expression/Homer | 1e-3 | -7.876e+00 | 0.0023 | 97.0 | 2.92% | 437.2 | 2.03% | motif file (matrix) |
pdf |
| 66 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.771e+00 | 0.0025 | 75.0 | 2.26% | 321.9 | 1.50% | motif file (matrix) |
pdf |
| 67 | 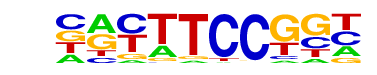 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-3 | -7.690e+00 | 0.0026 | 962.0 | 28.94% | 5672.5 | 26.37% | motif file (matrix) |
pdf |
| 68 |  | Dorsal(RHD)/Embryo-dl-ChIP-Seq(GSE65441)/Homer | 1e-3 | -7.521e+00 | 0.0031 | 157.0 | 4.72% | 776.1 | 3.61% | motif file (matrix) |
pdf |
| 69 |  | REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer | 1e-3 | -7.510e+00 | 0.0031 | 3.0 | 0.09% | 1.2 | 0.01% | motif file (matrix) |
pdf |
| 70 |  | Pax8(Paired,Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer | 1e-3 | -7.231e+00 | 0.0040 | 99.0 | 2.98% | 457.8 | 2.13% | motif file (matrix) |
pdf |
| 71 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-3 | -6.996e+00 | 0.0050 | 195.0 | 5.87% | 1005.2 | 4.67% | motif file (matrix) |
pdf |
| 72 |  | Unknown-ESC-element(?)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-2 | -6.463e+00 | 0.0084 | 142.0 | 4.27% | 711.3 | 3.31% | motif file (matrix) |
pdf |
| 73 |  | PAX5(Paired,Homeobox),condensed/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.449e+00 | 0.0084 | 65.0 | 1.96% | 284.7 | 1.32% | motif file (matrix) |
pdf |
| 74 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-2 | -6.228e+00 | 0.0102 | 51.0 | 1.53% | 213.8 | 0.99% | motif file (matrix) |
pdf |
| 75 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-2 | -6.228e+00 | 0.0102 | 51.0 | 1.53% | 213.8 | 0.99% | motif file (matrix) |
pdf |
| 76 |  | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-2 | -6.022e+00 | 0.0124 | 61.0 | 1.84% | 268.1 | 1.25% | motif file (matrix) |
pdf |
| 77 |  | CRE(bZIP)/Promoter/Homer | 1e-2 | -5.873e+00 | 0.0141 | 138.0 | 4.15% | 700.0 | 3.25% | motif file (matrix) |
pdf |
| 78 |  | RXR(NR),DR1/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.592e+00 | 0.0185 | 368.0 | 11.07% | 2078.8 | 9.66% | motif file (matrix) |
pdf |
| 79 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.286e+00 | 0.0248 | 332.0 | 9.99% | 1870.4 | 8.70% | motif file (matrix) |
pdf |
| 80 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-2 | -5.116e+00 | 0.0290 | 584.0 | 17.57% | 3429.8 | 15.94% | motif file (matrix) |
pdf |
| 81 |  | Zic(Zf)/Cerebellum-ZIC1.2-ChIP-Seq(GSE60731)/Homer | 1e-2 | -5.042e+00 | 0.0309 | 310.0 | 9.33% | 1745.9 | 8.12% | motif file (matrix) |
pdf |
| 82 |  | Srebp2(bHLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.777e+00 | 0.0397 | 55.0 | 1.65% | 252.5 | 1.17% | motif file (matrix) |
pdf |
| 83 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-2 | -4.747e+00 | 0.0405 | 27.0 | 0.81% | 105.3 | 0.49% | motif file (matrix) |
pdf |





































































































































































