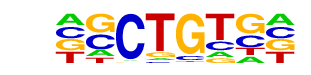
| p-value: | 1e-30 |
| log p-value: | -6.949e+01 |
| Information Content per bp: | 1.794 |
| Number of Target Sequences with motif | 493.0 |
| Percentage of Target Sequences with motif | 14.83% |
| Number of Background Sequences with motif | 1871.0 |
| Percentage of Background Sequences with motif | 8.70% |
| Average Position of motif in Targets | 385.7 +/- 309.9bp |
| Average Position of motif in Background | 259.1 +/- 209.3bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.10 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 1 |
| Score: | 0.72
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CACTGTGC
-GCTGTG- |
|

|
|
AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CACTGTGC----
--CTGTTCCTGG |
|


|
|
MET28/MA0332.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.68
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CACTGTGC
--CTGTGG |
|


|
|
MET28(MacIsaac)/Yeast
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CACTGTGC
--CTGTGG |
|


|
|
PB0199.1_Zfp161_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.68
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CACTGTGC---
NNGCNCTGCGCGGC |
|


|
|
HMRA1/MA0327.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CACTGTGC
-ATTGTGC |
|


|
|
MATA1(MacIsaac)/Yeast
| Match Rank: | 7 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CACTGTGC
-ATTGTGC |
|


|
|
CDC5(MYB)/Arabidopsis thaliana/AthaMap
| Match Rank: | 8 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CACTGTGC-
CGCGCTGAGCN |
|


|
|
CDC5/MA0579.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CACTGTGC-
CGCGCTGAGCN |
|


|
|
ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer
| Match Rank: | 10 |
| Score: | 0.66
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CACTGTGC-
WDNCTGGGCA |
|


|
|





















