| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | ABF1/SacCer-Promoters/Homer | 1e-226 | -5.216e+02 | 0.0000 | 1564.0 | 35.01% | 3133.7 | 15.42% | motif file (matrix) |
pdf |
| 2 |  | REB1/SacCer-Promoters/Homer | 1e-185 | -4.278e+02 | 0.0000 | 985.0 | 22.05% | 1620.5 | 7.98% | motif file (matrix) |
pdf |
| 3 |  | RLR1?/SacCer-Promoters/Homer | 1e-64 | -1.476e+02 | 0.0000 | 2115.0 | 47.35% | 7107.5 | 34.98% | motif file (matrix) |
pdf |
| 4 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-57 | -1.315e+02 | 0.0000 | 601.0 | 13.45% | 1363.7 | 6.71% | motif file (matrix) |
pdf |
| 5 |  | SeqBias: CG-repeat | 1e-46 | -1.075e+02 | 0.0000 | 799.0 | 17.89% | 2166.5 | 10.66% | motif file (matrix) |
pdf |
| 6 |  | Cbf1(bHLH)/Yeast-Cbf1-ChIP-Seq(GSE29506)/Homer | 1e-43 | -1.002e+02 | 0.0000 | 429.0 | 9.60% | 940.3 | 4.63% | motif file (matrix) |
pdf |
| 7 |  | bHLHE40(bHLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-43 | -9.948e+01 | 0.0000 | 441.0 | 9.87% | 981.1 | 4.83% | motif file (matrix) |
pdf |
| 8 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-38 | -8.811e+01 | 0.0000 | 412.0 | 9.22% | 936.0 | 4.61% | motif file (matrix) |
pdf |
| 9 |  | DPL-1(E2F)/cElegans-Adult-ChIP-Seq(modEncode)/Homer | 1e-38 | -8.786e+01 | 0.0000 | 1103.0 | 24.69% | 3454.5 | 17.00% | motif file (matrix) |
pdf |
| 10 |  | SFP1/SacCer-Promoters/Homer | 1e-37 | -8.591e+01 | 0.0000 | 659.0 | 14.75% | 1793.9 | 8.83% | motif file (matrix) |
pdf |
| 11 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-36 | -8.426e+01 | 0.0000 | 657.0 | 14.71% | 1796.7 | 8.84% | motif file (matrix) |
pdf |
| 12 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-34 | -7.987e+01 | 0.0000 | 486.0 | 10.88% | 1221.6 | 6.01% | motif file (matrix) |
pdf |
| 13 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-34 | -7.837e+01 | 0.0000 | 571.0 | 12.78% | 1525.6 | 7.51% | motif file (matrix) |
pdf |
| 14 |  | TATA-box/SacCer-Promoters/Homer | 1e-32 | -7.433e+01 | 0.0000 | 2123.0 | 47.53% | 7871.4 | 38.74% | motif file (matrix) |
pdf |
| 15 |  | E-box/Arabidopsis-Promoters/Homer | 1e-27 | -6.224e+01 | 0.0000 | 522.0 | 11.69% | 1451.8 | 7.14% | motif file (matrix) |
pdf |
| 16 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-25 | -5.870e+01 | 0.0000 | 430.0 | 9.63% | 1146.7 | 5.64% | motif file (matrix) |
pdf |
| 17 |  | Max(bHLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-24 | -5.755e+01 | 0.0000 | 590.0 | 13.21% | 1733.6 | 8.53% | motif file (matrix) |
pdf |
| 18 |  | E-box(bHLH)/Promoter/Homer | 1e-23 | -5.380e+01 | 0.0000 | 124.0 | 2.78% | 196.8 | 0.97% | motif file (matrix) |
pdf |
| 19 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-21 | -4.989e+01 | 0.0000 | 1366.0 | 30.58% | 4919.8 | 24.21% | motif file (matrix) |
pdf |
| 20 |  | SPCH(bHLH)/Seedling-SPCH-ChIP-Seq(GSE57497)/Homer | 1e-21 | -4.849e+01 | 0.0000 | 1113.0 | 24.92% | 3883.4 | 19.11% | motif file (matrix) |
pdf |
| 21 |  | FHY3(FAR1)/Arabidopsis-FHY3-ChIP-Seq(GSE30711)/Homer | 1e-20 | -4.626e+01 | 0.0000 | 426.0 | 9.54% | 1213.5 | 5.97% | motif file (matrix) |
pdf |
| 22 |  | PIF5ox(bHLH)/Arabidopsis-PIF5ox-ChIP-Seq(GSE35062)/Homer | 1e-20 | -4.609e+01 | 0.0000 | 876.0 | 19.61% | 2947.2 | 14.50% | motif file (matrix) |
pdf |
| 23 |  | PIF4(bHLH)/Seedling-PIF4-ChIP-Seq(GSE35315)/Homer | 1e-19 | -4.547e+01 | 0.0000 | 974.0 | 21.80% | 3350.4 | 16.49% | motif file (matrix) |
pdf |
| 24 |  | TOD6?/SacCer-Promoters/Homer | 1e-18 | -4.147e+01 | 0.0000 | 346.0 | 7.75% | 958.9 | 4.72% | motif file (matrix) |
pdf |
| 25 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-17 | -4.080e+01 | 0.0000 | 808.0 | 18.09% | 2733.3 | 13.45% | motif file (matrix) |
pdf |
| 26 |  | Egr2(Zf)/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-16 | -3.872e+01 | 0.0000 | 137.0 | 3.07% | 276.3 | 1.36% | motif file (matrix) |
pdf |
| 27 |  | NPAS2(bHLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-16 | -3.750e+01 | 0.0000 | 827.0 | 18.51% | 2848.7 | 14.02% | motif file (matrix) |
pdf |
| 28 |  | KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer | 1e-12 | -2.830e+01 | 0.0000 | 548.0 | 12.27% | 1839.4 | 9.05% | motif file (matrix) |
pdf |
| 29 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-11 | -2.680e+01 | 0.0000 | 794.0 | 17.77% | 2854.5 | 14.05% | motif file (matrix) |
pdf |
| 30 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-10 | -2.520e+01 | 0.0000 | 150.0 | 3.36% | 376.9 | 1.85% | motif file (matrix) |
pdf |
| 31 |  | Arnt:Ahr(bHLH)/MCF7-Arnt-ChIP-Seq(Lo_et_al.)/Homer | 1e-10 | -2.486e+01 | 0.0000 | 1056.0 | 23.64% | 3981.8 | 19.60% | motif file (matrix) |
pdf |
| 32 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-10 | -2.455e+01 | 0.0000 | 1689.0 | 37.81% | 6727.1 | 33.11% | motif file (matrix) |
pdf |
| 33 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-10 | -2.432e+01 | 0.0000 | 1427.0 | 31.95% | 5585.4 | 27.49% | motif file (matrix) |
pdf |
| 34 |  | KLF10(Zf)/HEK293-KLF10.GFP-ChIP-Seq(GSE58341)/Homer | 1e-10 | -2.411e+01 | 0.0000 | 242.0 | 5.42% | 707.9 | 3.48% | motif file (matrix) |
pdf |
| 35 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-10 | -2.347e+01 | 0.0000 | 317.0 | 7.10% | 992.4 | 4.88% | motif file (matrix) |
pdf |
| 36 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-10 | -2.322e+01 | 0.0000 | 157.0 | 3.51% | 411.9 | 2.03% | motif file (matrix) |
pdf |
| 37 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-10 | -2.320e+01 | 0.0000 | 330.0 | 7.39% | 1044.2 | 5.14% | motif file (matrix) |
pdf |
| 38 |  | SUT1?/SacCer-Promoters/Homer | 1e-9 | -2.251e+01 | 0.0000 | 3023.0 | 67.67% | 12835.2 | 63.17% | motif file (matrix) |
pdf |
| 39 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-8 | -2.069e+01 | 0.0000 | 289.0 | 6.47% | 912.8 | 4.49% | motif file (matrix) |
pdf |
| 40 |  | ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer | 1e-8 | -2.063e+01 | 0.0000 | 315.0 | 7.05% | 1012.7 | 4.98% | motif file (matrix) |
pdf |
| 41 |  | GAGA-repeat/Arabidopsis-Promoters/Homer | 1e-8 | -1.893e+01 | 0.0000 | 534.0 | 11.95% | 1903.3 | 9.37% | motif file (matrix) |
pdf |
| 42 |  | SeqBias: TA-repeat | 1e-8 | -1.848e+01 | 0.0000 | 3975.0 | 88.99% | 17505.2 | 86.15% | motif file (matrix) |
pdf |
| 43 |  | Klf9(Zf)/GBM-Klf9-ChIP-Seq(GSE62211)/Homer | 1e-7 | -1.786e+01 | 0.0000 | 127.0 | 2.84% | 340.5 | 1.68% | motif file (matrix) |
pdf |
| 44 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-7 | -1.668e+01 | 0.0000 | 104.0 | 2.33% | 268.5 | 1.32% | motif file (matrix) |
pdf |
| 45 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-7 | -1.624e+01 | 0.0000 | 456.0 | 10.21% | 1625.6 | 8.00% | motif file (matrix) |
pdf |
| 46 |  | HIF-1a(bHLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer | 1e-6 | -1.559e+01 | 0.0000 | 493.0 | 11.04% | 1786.1 | 8.79% | motif file (matrix) |
pdf |
| 47 |  | HIF2a(bHLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer | 1e-6 | -1.547e+01 | 0.0000 | 744.0 | 16.66% | 2834.3 | 13.95% | motif file (matrix) |
pdf |
| 48 | 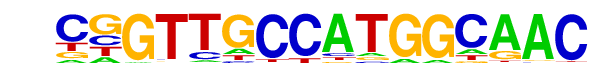 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-6 | -1.507e+01 | 0.0000 | 96.0 | 2.15% | 250.4 | 1.23% | motif file (matrix) |
pdf |
| 49 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-5 | -1.376e+01 | 0.0000 | 130.0 | 2.91% | 379.9 | 1.87% | motif file (matrix) |
pdf |
| 50 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-5 | -1.299e+01 | 0.0000 | 45.0 | 1.01% | 94.9 | 0.47% | motif file (matrix) |
pdf |
| 51 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-5 | -1.243e+01 | 0.0000 | 1457.0 | 32.62% | 6000.7 | 29.53% | motif file (matrix) |
pdf |
| 52 |  | TATA-box/Drosophila-Promoters/Homer | 1e-5 | -1.222e+01 | 0.0000 | 232.0 | 5.19% | 782.3 | 3.85% | motif file (matrix) |
pdf |
| 53 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-5 | -1.221e+01 | 0.0000 | 194.0 | 4.34% | 634.1 | 3.12% | motif file (matrix) |
pdf |
| 54 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-5 | -1.191e+01 | 0.0000 | 607.0 | 13.59% | 2327.7 | 11.46% | motif file (matrix) |
pdf |
| 55 |  | Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer | 1e-5 | -1.188e+01 | 0.0000 | 97.0 | 2.17% | 274.8 | 1.35% | motif file (matrix) |
pdf |
| 56 |  | NRF(NRF)/Promoter/Homer | 1e-4 | -1.144e+01 | 0.0001 | 95.0 | 2.13% | 270.2 | 1.33% | motif file (matrix) |
pdf |
| 57 |  | SpiB(ETS)/OCILY3-SPIB-ChIP-Seq(GSE56857)/Homer | 1e-4 | -1.133e+01 | 0.0001 | 300.0 | 6.72% | 1065.6 | 5.24% | motif file (matrix) |
pdf |
| 58 |  | E2F(E2F)/Hela-CellCycle-Expression/Homer | 1e-4 | -9.700e+00 | 0.0004 | 132.0 | 2.96% | 422.1 | 2.08% | motif file (matrix) |
pdf |
| 59 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-3 | -8.953e+00 | 0.0008 | 73.0 | 1.63% | 209.6 | 1.03% | motif file (matrix) |
pdf |
| 60 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-3 | -8.953e+00 | 0.0008 | 73.0 | 1.63% | 209.6 | 1.03% | motif file (matrix) |
pdf |
| 61 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-3 | -8.498e+00 | 0.0013 | 775.0 | 17.35% | 3129.1 | 15.40% | motif file (matrix) |
pdf |
| 62 |  | X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer | 1e-3 | -8.420e+00 | 0.0014 | 136.0 | 3.04% | 451.6 | 2.22% | motif file (matrix) |
pdf |
| 63 |  | Pax8(Paired,Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer | 1e-3 | -8.351e+00 | 0.0015 | 134.0 | 3.00% | 444.7 | 2.19% | motif file (matrix) |
pdf |
| 64 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.100e+00 | 0.0018 | 288.0 | 6.45% | 1068.1 | 5.26% | motif file (matrix) |
pdf |
| 65 |  | Trl(Zf)/S2-GAGAfactor-ChIP-Seq(GSE40646)/Homer | 1e-3 | -8.069e+00 | 0.0019 | 1708.0 | 38.24% | 7266.5 | 35.76% | motif file (matrix) |
pdf |
| 66 |  | CRE(bZIP)/Promoter/Homer | 1e-3 | -7.578e+00 | 0.0030 | 185.0 | 4.14% | 656.0 | 3.23% | motif file (matrix) |
pdf |
| 67 |  | ZBTB33(Zf)/GM12878-ZBTB33-ChIP-Seq(GSE32465)/Homer | 1e-3 | -7.531e+00 | 0.0031 | 52.0 | 1.16% | 144.5 | 0.71% | motif file (matrix) |
pdf |
| 68 |  | PAX5(Paired,Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.143e+00 | 0.0122 | 209.0 | 4.68% | 777.5 | 3.83% | motif file (matrix) |
pdf |
| 69 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer | 1e-2 | -6.135e+00 | 0.0122 | 44.0 | 0.99% | 125.7 | 0.62% | motif file (matrix) |
pdf |
| 70 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.037e+00 | 0.0132 | 1017.0 | 22.77% | 4271.9 | 21.02% | motif file (matrix) |
pdf |
| 71 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.884e+00 | 0.0152 | 41.0 | 0.92% | 116.3 | 0.57% | motif file (matrix) |
pdf |
| 72 |  | RXR(NR),DR1/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.651e+00 | 0.0189 | 516.0 | 11.55% | 2092.8 | 10.30% | motif file (matrix) |
pdf |
| 73 |  | PAX5(Paired,Homeobox),condensed/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.438e+00 | 0.0231 | 77.0 | 1.72% | 255.0 | 1.26% | motif file (matrix) |
pdf |
| 74 |  | Dorsal(RHD)/Embryo-dl-ChIP-Seq(GSE65441)/Homer | 1e-2 | -5.168e+00 | 0.0298 | 212.0 | 4.75% | 808.1 | 3.98% | motif file (matrix) |
pdf |
| 75 |  | PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer | 1e-2 | -5.062e+00 | 0.0327 | 33.0 | 0.74% | 93.7 | 0.46% | motif file (matrix) |
pdf |
| 76 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.001e+00 | 0.0343 | 832.0 | 18.63% | 3496.3 | 17.21% | motif file (matrix) |
pdf |
| 77 |  | ZNF692(Zf)/HEK293-ZNF692.GFP-ChIP-Seq(GSE58341)/Homer | 1e-2 | -4.988e+00 | 0.0343 | 55.0 | 1.23% | 175.4 | 0.86% | motif file (matrix) |
pdf |
| 78 |  | Unknown-ESC-element(?)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-2 | -4.985e+00 | 0.0343 | 178.0 | 3.98% | 670.7 | 3.30% | motif file (matrix) |
pdf |
| 79 |  | ZNF519(Zf)/HEK293-ZNF519.GFP-ChIP-Seq(GSE58341)/Homer | 1e-2 | -4.800e+00 | 0.0403 | 49.0 | 1.10% | 154.2 | 0.76% | motif file (matrix) |
pdf |
| 80 |  | GEI-11(Myb?)/cElegans-L4-GEI11-ChIP-Seq(modEncode)/Homer | 1e-2 | -4.771e+00 | 0.0410 | 42.0 | 0.94% | 128.4 | 0.63% | motif file (matrix) |
pdf |
| 81 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-2 | -4.687e+00 | 0.0440 | 350.0 | 7.84% | 1405.3 | 6.92% | motif file (matrix) |
pdf |

































































































































































