| p-value: | 1e-17 |
| log p-value: | -3.973e+01 |
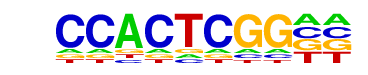
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 80.0 |
| Percentage of Target Sequences with motif | 1.85% |
| Number of Background Sequences with motif | 126.4 |
| Percentage of Background Sequences with motif | 0.59% |
| Average Position of motif in Targets | 347.5 +/- 259.8bp |
| Average Position of motif in Background | 244.7 +/- 200.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Bapx1(Homeobox)/VertebralCol-Bapx1-ChIP-Seq(GSE36672)/Homer
| Match Rank: | 1 |
| Score: | 0.75
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CCACTCGG
MRSCACTYAA |
|


|
|
PB0114.1_Egr1_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.74
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------CCACTCGG--
NNAGTCCCACTCNNNN |
|


|
|
BHLH112/MA0961.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.74
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCACTCGG
GCCACTTGC |
|


|
|
tin/MA0247.2/Jaspar
| Match Rank: | 4 |
| Score: | 0.73
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCACTCGG--
CCACTTGANA |
|

|
|
z/dmmpmm(Down)/fly
| Match Rank: | 5 |
| Score: | 0.72
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CCACTCGG
-CACTCA- |
|


|
|
z/dmmpmm(Bigfoot)/fly
| Match Rank: | 6 |
| Score: | 0.70
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CCACTCGG
-CACTCA- |
|


|
|
NFY(CCAAT)/Promoter/Homer
| Match Rank: | 7 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCACTCGG
AGCCAATCGG |
|


|
|
Nkx2.2(Homeobox)/NPC-Nkx2.2-ChIP-Seq(GSE61673)/Homer
| Match Rank: | 8 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCACTCGG-
NSCACTYVAV |
|


|
|
Nkx2.5(Homeobox)/HL1-Nkx2.5.biotin-ChIP-Seq(GSE21529)/Homer
| Match Rank: | 9 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCACTCGG
AASCACTCAA |
|


|
|
CMTA3/MA0970.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCACTCGG
NNNACGCGG |
|


|
|





















