| p-value: | 1e-12 |
| log p-value: | -2.925e+01 |
| Information Content per bp: | 1.930 |
| Number of Target Sequences with motif | 116.0 |
| Percentage of Target Sequences with motif | 2.72% |
| Number of Background Sequences with motif | 269.3 |
| Percentage of Background Sequences with motif | 1.28% |
| Average Position of motif in Targets | 474.4 +/- 302.2bp |
| Average Position of motif in Background | 279.1 +/- 201.9bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.11 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
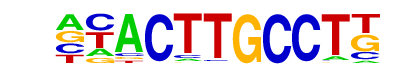
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 1 |
| Score: | 0.87
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CACTTGCC--
NNACTTGCCTT |
|

|
|
INO2/INO2_YPD/1-INO4,2-INO2(Harbison)/Yeast
| Match Rank: | 2 |
| Score: | 0.87
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CACTTGCC
CACATGC- |
|


|
|
IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer
| Match Rank: | 3 |
| Score: | 0.81
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CACTTGCC
CACGTGCC |
|


|
|
HBI1/MA1025.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.80
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CACTTGCC--
NGCACGTGCNNN |
|


|
|
BHLH112/MA0961.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.79
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CACTTGCC
GCCACTTGC- |
|


|
|
PIF4/MA0561.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.78
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CACTTGCC
CACGTGGC |
|


|
|
BHLH104/MA0960.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.78
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CACTTGCC
GGCACGTGCC |
|


|
|
HEY1/MA0823.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.78
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CACTTGCC
GACACGTGCC |
|


|
|
PL0016.1_ref-1/Jaspar
| Match Rank: | 9 |
| Score: | 0.78
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----CACTTGCC----
TNNGGCACGTGCCNTNN |
|


|
|
sna/dmmpmm(Papatsenko)/fly
| Match Rank: | 10 |
| Score: | 0.78
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CACTTGCC
CCACCTGC- |
|


|
|





















