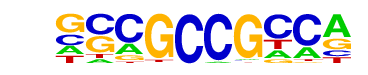
| p-value: | 1e-30 |
| log p-value: | -6.997e+01 |
| Information Content per bp: | 1.762 |
| Number of Target Sequences with motif | 496.0 |
| Percentage of Target Sequences with motif | 11.29% |
| Number of Background Sequences with motif | 1238.0 |
| Percentage of Background Sequences with motif | 6.56% |
| Average Position of motif in Targets | 499.4 +/- 354.1bp |
| Average Position of motif in Background | 324.3 +/- 276.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.13 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
ERF6/MA1006.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.86
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGGCCGGC--
CTGCCGGCGT |
|


|
|
YLL054C/MA0429.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.86
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CGGCCGGC
TCGGCCG-- |
|


|
|
RDS1(MacIsaac)/Yeast
| Match Rank: | 3 |
| Score: | 0.86
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGGCCGGC
CGGCCG-- |
|


|
|
RDS1/MA0361.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.85
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CGGCCGGC
NCGGCCG-- |
|


|
|
ERF105/MA1000.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.83
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CGGCCGGC-
-TGCCGGCG |
|


|
|
ERF098/MA0999.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.81
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGGCCGGC
CCGCCGCC |
|


|
|
ERF13/MA1004.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.80
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CGGCCGGC-
-CGCCGCCA |
|


|
|
ERF4/MA0992.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.80
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGGCCGGC
CCGCCGCC |
|


|
|
ERF112/MA1002.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.79
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CGGCCGGC-
GCCGCCGCCA |
|

|
|
ERF096/MA0998.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.79
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGGCCGGC--
CCGCCGCCAT |
|


|
|





















