| p-value: | 1e-14 |
| log p-value: | -3.406e+01 |
| Information Content per bp: | 1.929 |
| Number of Target Sequences with motif | 186.0 |
| Percentage of Target Sequences with motif | 4.68% |
| Number of Background Sequences with motif | 571.3 |
| Percentage of Background Sequences with motif | 2.49% |
| Average Position of motif in Targets | 292.8 +/- 218.1bp |
| Average Position of motif in Background | 261.0 +/- 182.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.01 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
NRG1/NRG1_H2O2Hi/[](Harbison)/Yeast
| Match Rank: | 1 |
| Score: | 0.77
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AAGGGCCC
-AGGGTCC |
|


|
|
NRG1(MacIsaac)/Yeast
| Match Rank: | 2 |
| Score: | 0.74
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AAGGGCCC
-AGGGTCC |
|


|
|
NRG1/MA0347.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.74
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------AAGGGCCC------
CTAGATCAGGGTCCATCGCA |
|


|
|
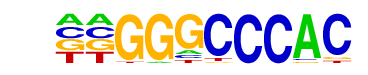
OsI_08196/MA1050.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.73
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AAGGGCCC--
--GGGCCCAC |
|

|
|
OJ1581_H09.2/MA1031.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.72
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAGGGCCC--
GTGGGCCCAC |
|


|
|
TCP23/MA1066.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.72
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AAGGGCCC--
--GGGCCCAC |
|


|
|
TCP15/MA1062.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.71
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AAGGGCCC--
--GGGCCCAC |
|


|
|
MSN2(MacIsaac)/Yeast
| Match Rank: | 8 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAGGGCCC
AAGGGGC- |
|


|
|
PCF5(TCP)/Oryza sativa/AthaMap
| Match Rank: | 9 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AAGGGCCC--
NNGGGACCAC |
|


|
|
TCP20/MA1065.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.68
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AAGGGCCC----
--GGGCCCACCA |
|


|
|





















