| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.72 |
RXR(NR),DR1/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer |
 |
0.78 |
>HES7_1HES7_jolma_DBD_M319
|
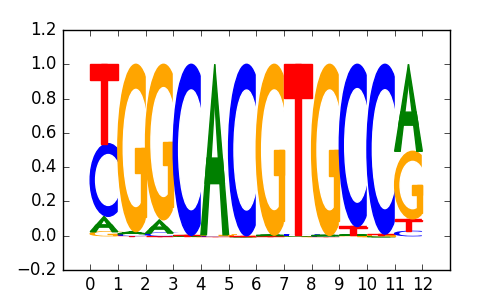 |
0.78 |
M5021_1.02.txt |
 |
| 0.7 |
BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer |
 |
0.76 |
>HES5_1HES5_jolma_DBD_M318
|
 |
0.78 |
HES7 |
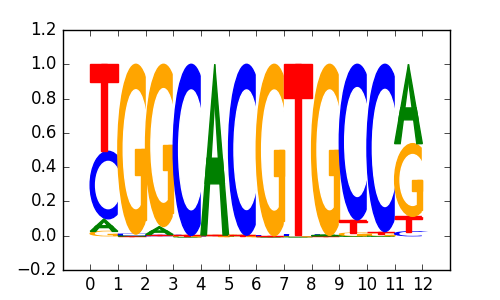 |
| 0.7 |
CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer |
 |
0.74 |
>CTCFL_disc1 CTCFL_K562_encode-Myers_seq_hsa_v041610.1-SC-98982_r1:MDscan#2#Intergenic
|
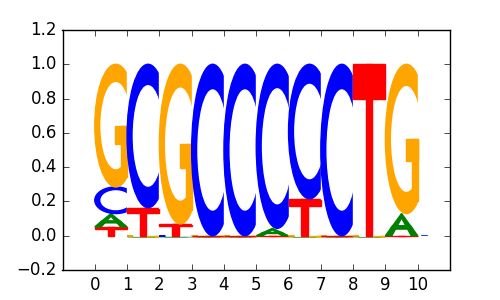 |
0.77 |
M5016_1.02.txt |
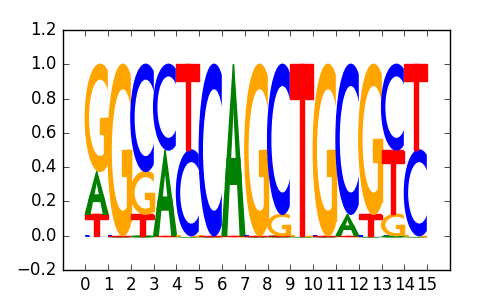 |
| 0.7 |
TR4(NR),DR1/Hela-TR4-ChIP-Seq(GSE24685)/Homer |
 |
0.74 |
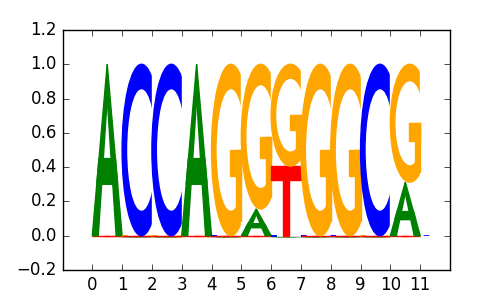
>CTCF_disc2 CTCF_A549_encode-Myers_seq_hsa_DEX-100nM_r1:Trawler#1#Intergenic
|
 |
0.76 |
HES5 |
 |
| 0.69 |
HNF4a(NR),DR1/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer |
 |
0.73 |
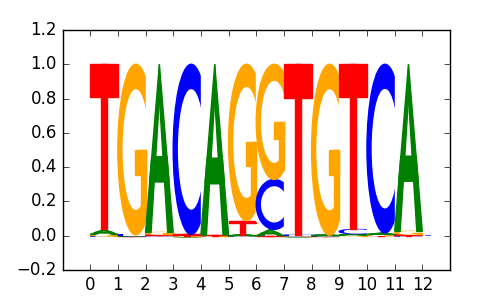
>PKNOX2_2PKNOX2_jolma_DBD_M172
|
 |
0.76 |
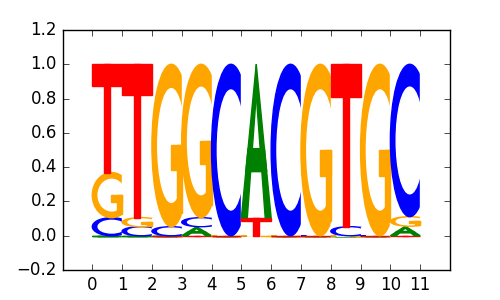
M5022_1.02.txt |
 |
| 0.69 |
KLF6(Zf)/PDAC-KLF6-ChIP-Seq(GSE64557)/Homer |
 |
0.73 |
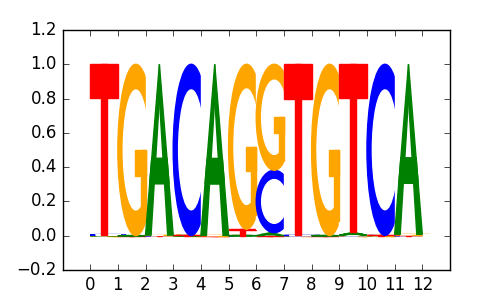
>PKNOX2_3Pknox2_jolma_DBD_M173
|
 |
0.76 |
HES5 |
 |
| 0.69 |
Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer |
 |
0.73 |
>TFAP2_disc2 TFAP2A_HeLa-S3_encode-Snyder_seq_hsa_r1:MEME#2#Intergenic
|
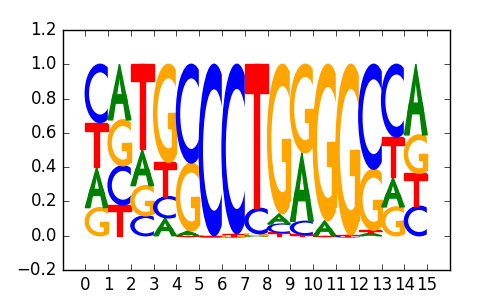 |
0.74 |
M1655_1.02.txt |
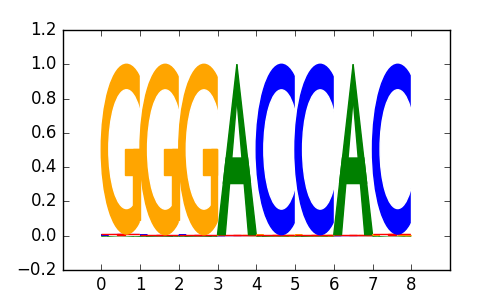 |
| 0.69 |
AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer |
 |
0.72 |
>PKNOX1_2PKNOX1_jolma_DBD_M171
|
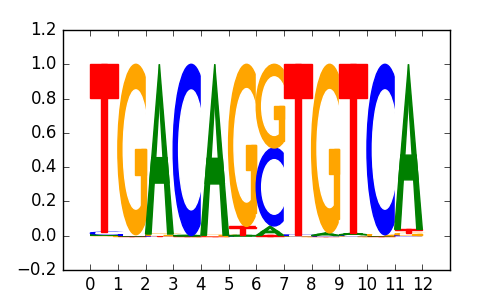 |
0.74 |
M0159_1.02.txt |
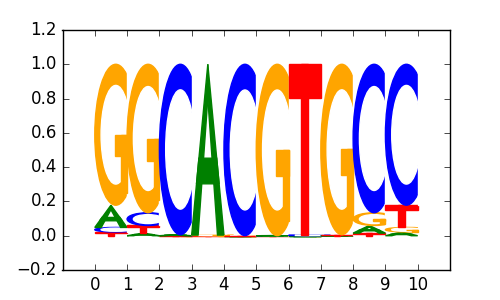 |
| 0.69 |
PPARE(NR),DR1/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer |
 |
0.72 |
>TCF3_3E12_transfac_M00693
|
 |
0.74 |
M5025_1.02.txt |
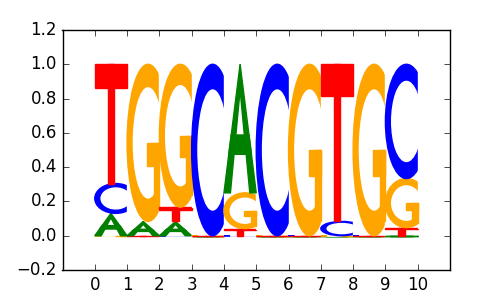 |
| 0.69 |
RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer |
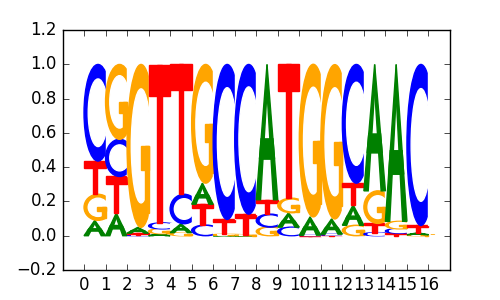 |
0.71 |
>HNF4_known7HNF4_4COUP-direct-repeat-1_transfac_M00765
|
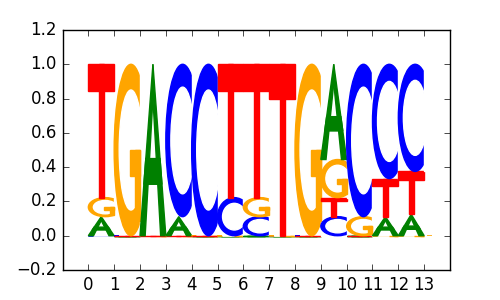 |
0.74 |
M2380_1.02.txt |
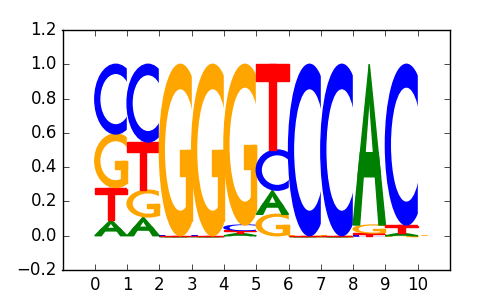 |

































