| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.76 |
BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer |
 |
0.81 |
>CTCFL_disc1 CTCFL_K562_encode-Myers_seq_hsa_v041610.1-SC-98982_r1:MDscan#2#Intergenic
|
 |
0.8 |
INSM1 |
 |
| 0.76 |
CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski_et_al.)/Homer |
 |
0.8 |
>ZBTB7A_disc1 ZBTB7A_K562_encode-Myers_seq_hsa_v041610.1-SC-34508_r1:MDscan#2#Intergenic
|
 |
0.78 |
PRDM4 |
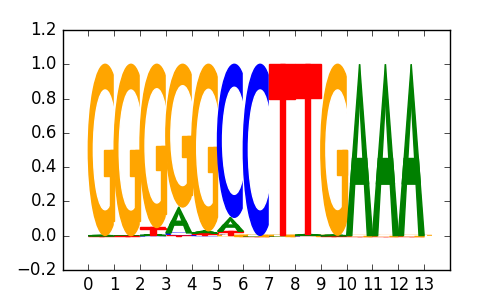 |
| 0.73 |
ZNF16(Zf)/HEK293-ZNF16.GFP-ChIP-Seq(GSE58341)/Homer |
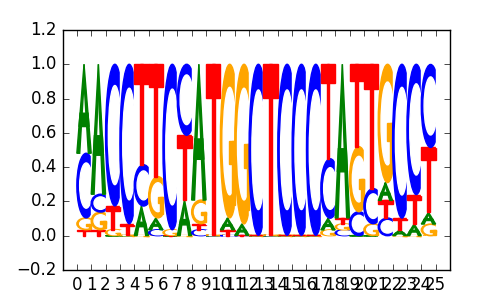 |
0.8 |
>CTCF_disc2 CTCF_A549_encode-Myers_seq_hsa_DEX-100nM_r1:Trawler#1#Intergenic
|
 |
0.78 |
M6113_1.02.txt |
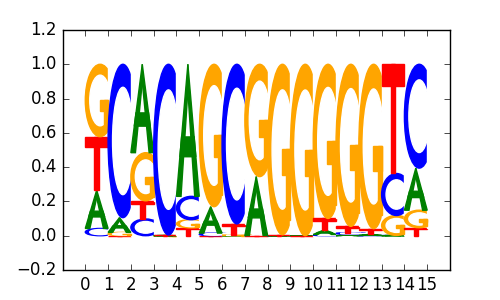 |
| 0.71 |
ZSCAN22(Zf)/HEK293-ZSCAN22.GFP-ChIP-Seq(GSE58341)/Homer |
 |
0.78 |
>PRDM4_1PRDM4_jolma_full_M742
|
 |
0.77 |
ZIC1 |
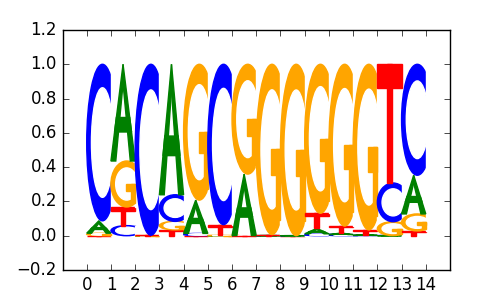 |
| 0.69 |
E2A(bHLH),near_PU.1/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer |
 |
0.78 |
>ZIC3_4Zic3_jolma_DBD_M57
|
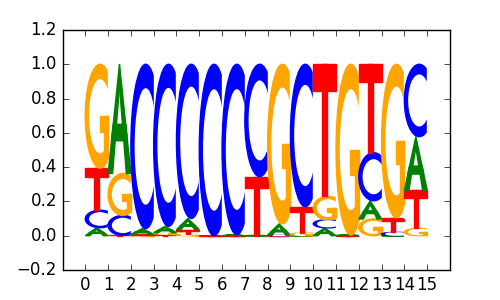 |
0.77 |
PLAGL1 |
 |
| 0.68 |
Zic3(Zf)/mES-Zic3-ChIP-Seq(GSE37889)/Homer |
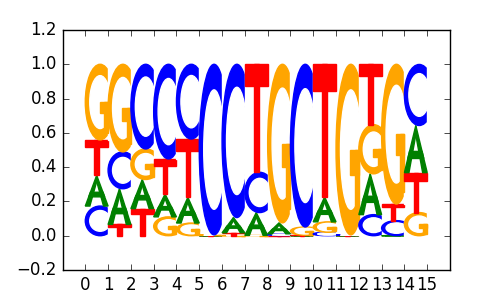 |
0.77 |
>ZIC1_3ZIC1_jolma_full_M40
|
 |
0.77 |
M6062_1.02.txt |
 |
| 0.68 |
NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer |
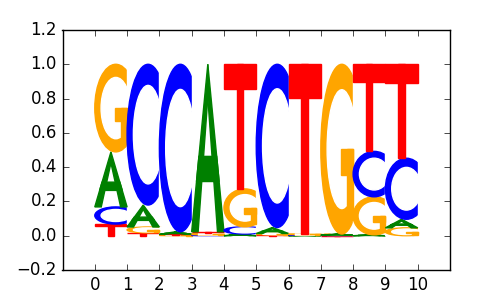 |
0.77 |
>PKNOX2_3Pknox2_jolma_DBD_M173
|
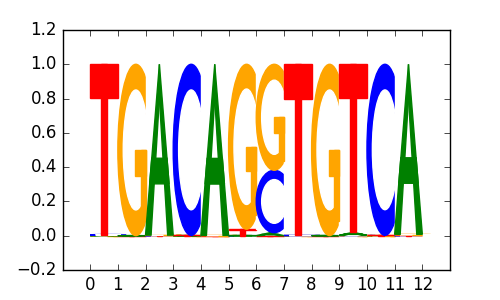 |
0.77 |
CTCFL |
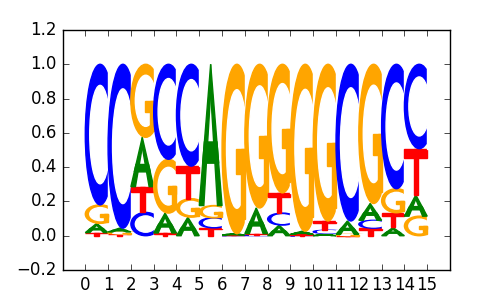 |
| 0.68 |
Sp1(Zf)/Promoter/Homer |
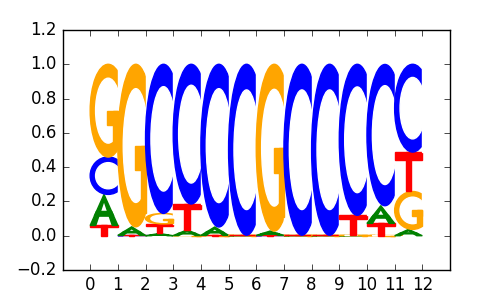 |
0.77 |
>PKNOX2_2PKNOX2_jolma_DBD_M172
|
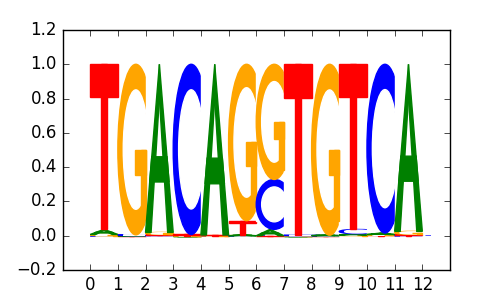 |
0.77 |
PKNOX2 |
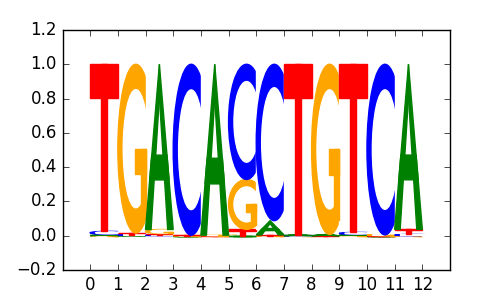 |
| 0.67 |
Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer |
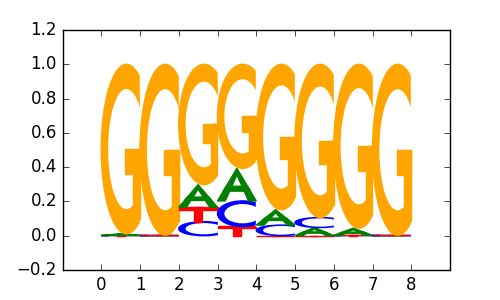 |
0.76 |
>ZIC3_3ZIC3_jolma_full_M41
|
 |
0.76 |
M2095_1.02.txt |
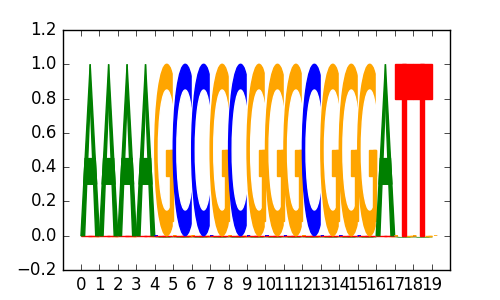 |
| 0.67 |
Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer |
 |
0.75 |
>PKNOX1_2PKNOX1_jolma_DBD_M171
|
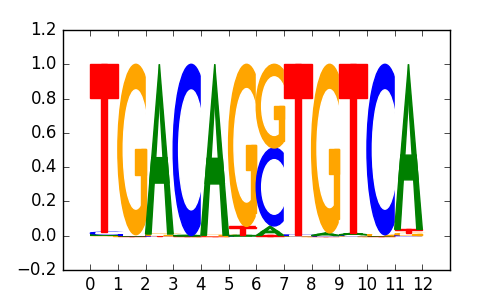 |
0.76 |
M5182_1.02.txt |
 |