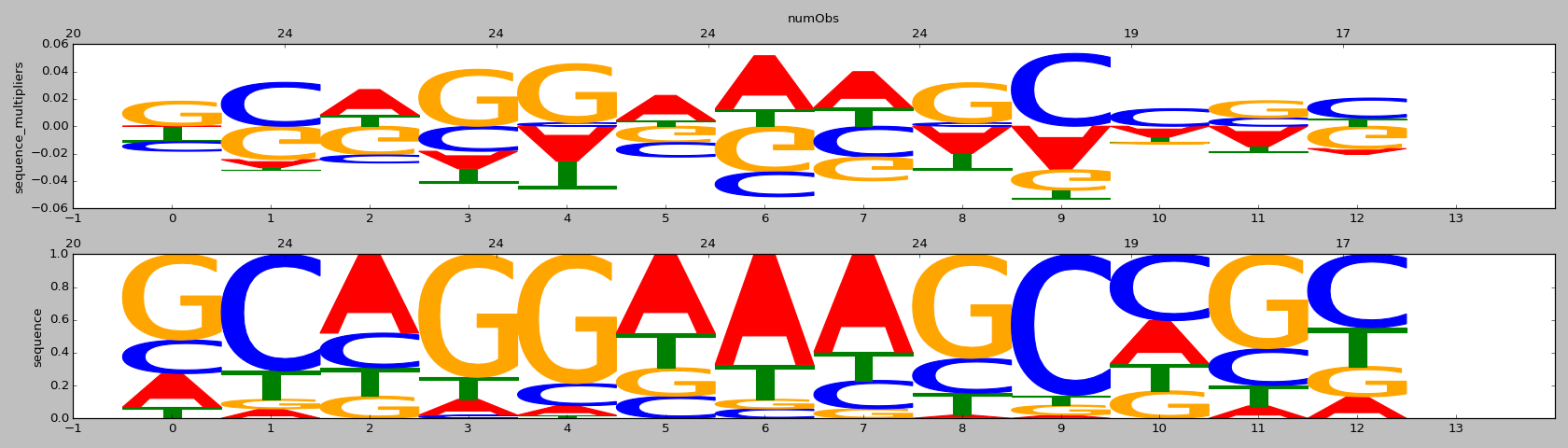
| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.79 |
TEAD2(TEA)/Py2T-Tead2-ChIP-Seq(GSE55709)/Homer |
 |
0.79 |
>FLI1_4FLI1_jolma_full_M115
|
 |
0.79 |
FLI1 |
 |
| 0.75 |
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer |
 |
0.79 |
>ERG_4ERG_jolma_full_M97
|
 |
0.79 |
ERG |
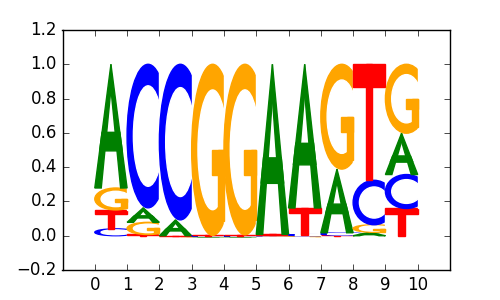 |
| 0.73 |
TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer |
 |
0.78 |
>ERG_2ERG_jolma_DBD_M95
|
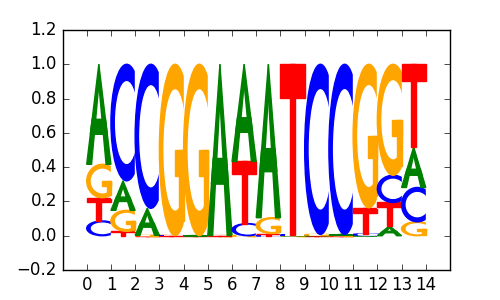 |
0.78 |
ERG |
 |
| 0.73 |
EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer |
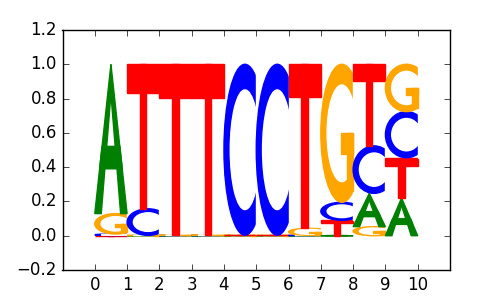 |
0.78 |
>BCL6B_2BCL6B_jolma_DBD_M1
|
 |
0.78 |
M5189_1.02.txt |
 |
| 0.73 |
EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer |
 |
0.77 |
>FLI1_2FLI1_jolma_DBD_M113
|
 |
0.78 |
BCL6B |
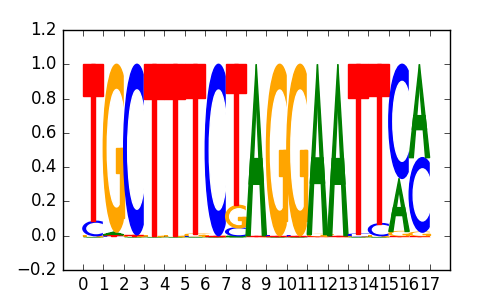 |
| 0.71 |
NFkB-p65-Rel(RHD)/ThioMac-LPS-Expression(GSE23622)/Homer |
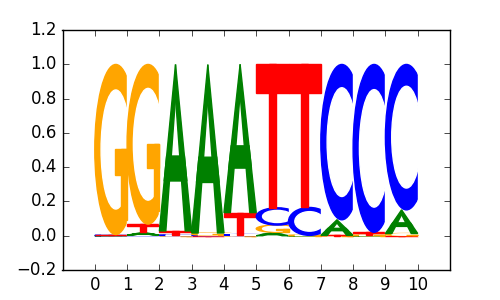 |
0.75 |
>BDP1_disc1 GTF3C2_K562_encode-Snyder_seq_hsa_r1:MEME#3#Intergenic
|
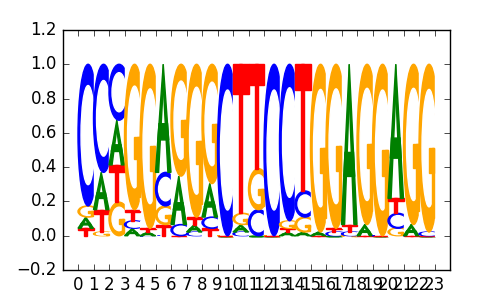 |
0.77 |
FLI1 |
 |
| 0.71 |
NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer |
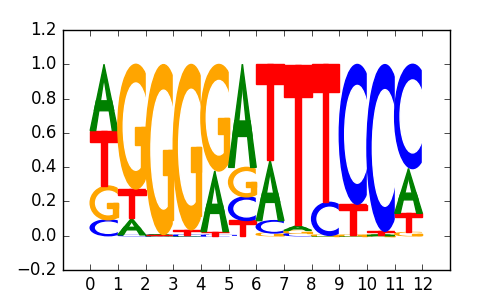 |
0.74 |
>NFKB_known9RELA_2RELA_jaspar_MA0107.1
|
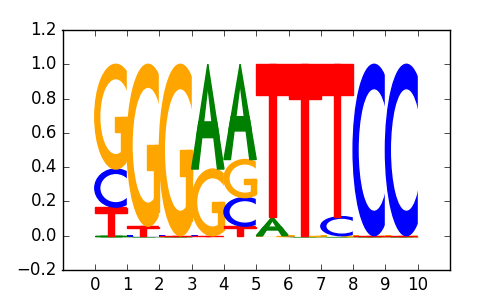 |
0.76 |
RELA |
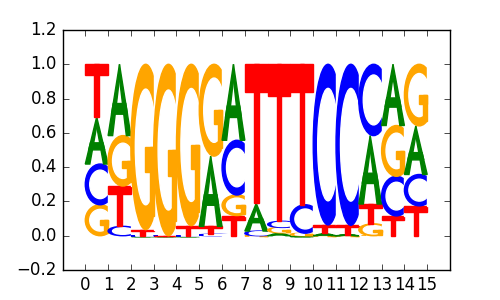 |
| 0.7 |
REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer |
 |
0.74 |
>TEAD1_2TEAD1_jaspar_MA0090.1
|
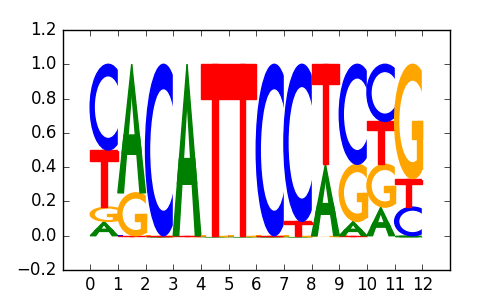 |
0.76 |
RELA |
 |
| 0.69 |
Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer |
 |
0.74 |
>TEAD3_2TEAD3_jolma_DBD_M289
|
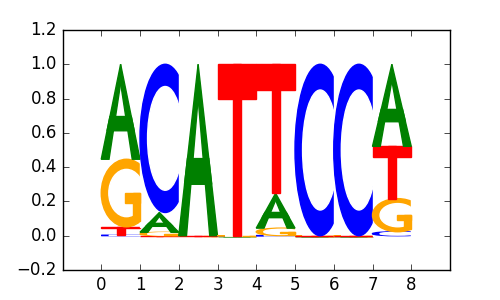 |
0.75 |
M1855_1.02.txt |
 |
| 0.69 |
ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer |
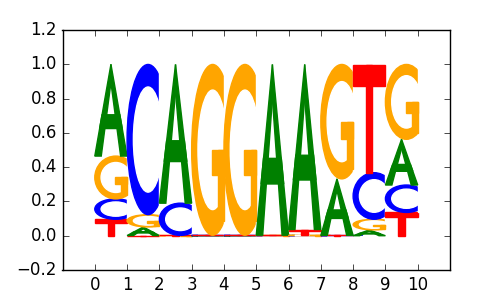 |
0.74 |
>NFKB_known2RELA_1NF-kappaB-(p65)_transfac_M00052
|
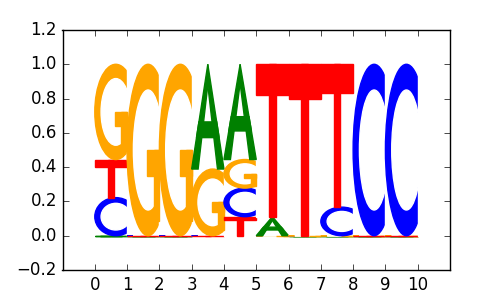 |
0.75 |
RELA |
 |