| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
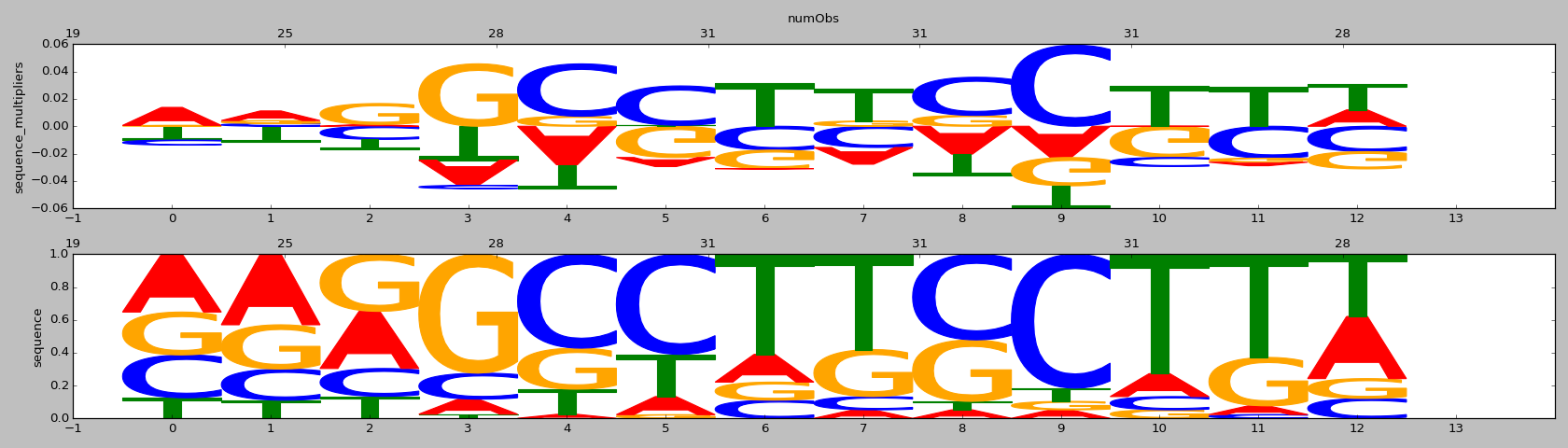
| 0.71 |
ZNF528(Zf)/HEK293-ZNF528.GFP-ChIP-Seq(GSE58341)/Homer |
 |
0.79 |
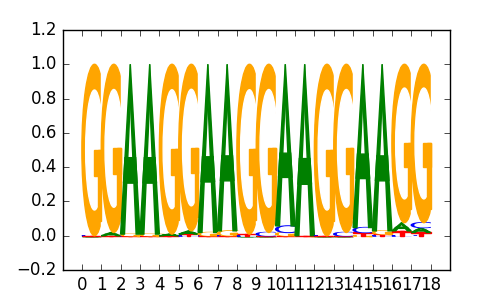
>EWSR1::FLI1_1EWSR1-FLI1_jaspar_MA0149.1
|
 |
0.76 |
IRF5 |
 |
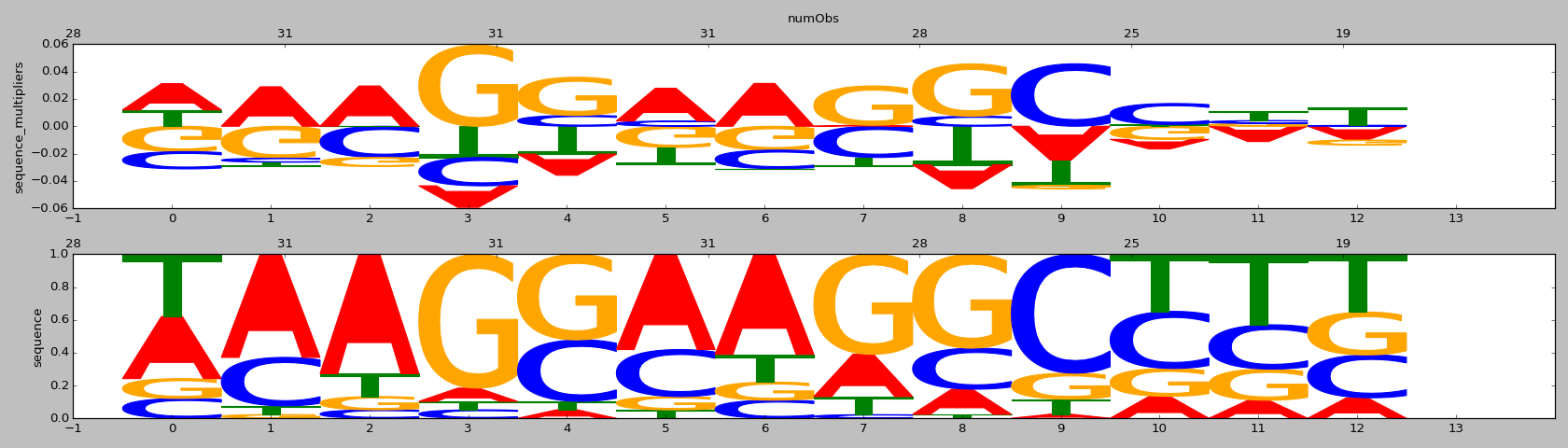
| 0.7 |
ZNF16(Zf)/HEK293-ZNF16.GFP-ChIP-Seq(GSE58341)/Homer |
 |
0.74 |
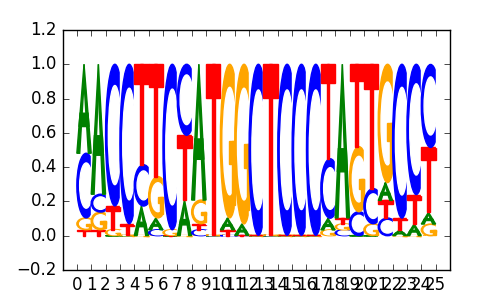
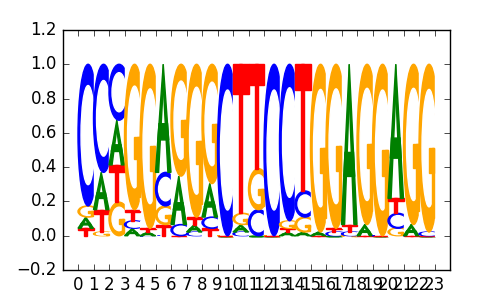
>BDP1_disc1 GTF3C2_K562_encode-Snyder_seq_hsa_r1:MEME#3#Intergenic
|
 |
0.73 |
M5189_1.02.txt |
 |
| 0.69 |
Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer |
 |
0.74 |
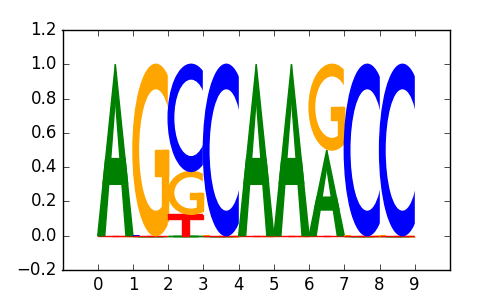
>GATA_disc5 GATA3_T-47D_encode-Myers_seq_hsa_DMSO-0.02pct-v041610.2-SC-268_r1:Trawler#2#Intergenic
|
 |
0.72 |
M2109_1.02.txt |
 |
| 0.68 |
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer |
 |
0.7 |
>ELF2_1NERF1a_transfac_M00531
|
 |
0.72 |
M4768_1.02.txt |
 |
| 0.68 |
TEAD2(TEA)/Py2T-Tead2-ChIP-Seq(GSE55709)/Homer |
 |
0.69 |
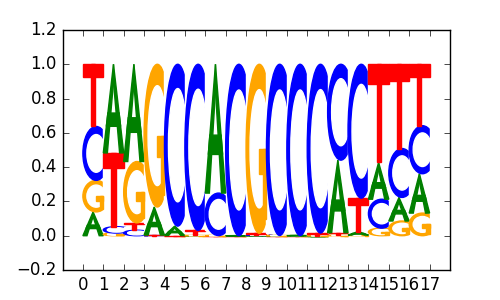
>SP4_2SP4_jolma_full_M31
|
 |
0.72 |
EHF |
 |
| 0.67 |
ETS:E-box(ETS,bHLH)/HPC7-Scl-ChIP-Seq(GSE22178)/Homer |
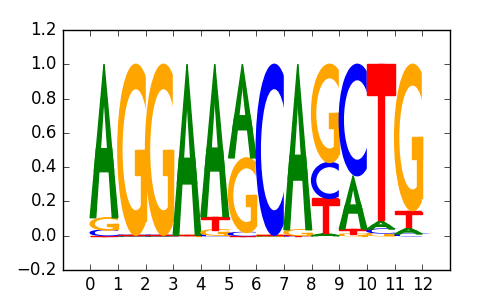 |
0.69 |
>EP300_disc10 EP300_HeLa-S3_encode-Snyder_seq_hsa_IgG-rab-N-15_r1:AlignACE#3#Intergenic
|
 |
0.7 |
M2431_1.02.txt |
 |
| 0.67 |
ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer |
 |
0.69 |
>ETS_known5ETS2_1c-Ets-2_transfac_M00340
|
 |
0.7 |
M4324_1.02.txt |
 |
| 0.67 |
ELF3(ETS)/PDAC-ELF3-ChIP-Seq(GSE64557)/Homer |
 |
0.69 |
>STAT_disc6 STAT3_MCF10A-Er-Src_encode-Snyder_seq_hsa_EtOH-0.01pct-4hr_r1:Trawler#3#Intergenic
|
 |
0.69 |
M2110_1.02.txt |
 |
| 0.67 |
PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer |
 |
0.69 |
>HIC1_4Hic1_jolma_DBD_M19
|
 |
0.69 |
SP4 |
 |
| 0.67 |
ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer |
 |
0.68 |
>IRF_known14IRF3_2IRF3_jolma_full_M147
|
 |
0.69 |
M6022_1.02.txt |
 |