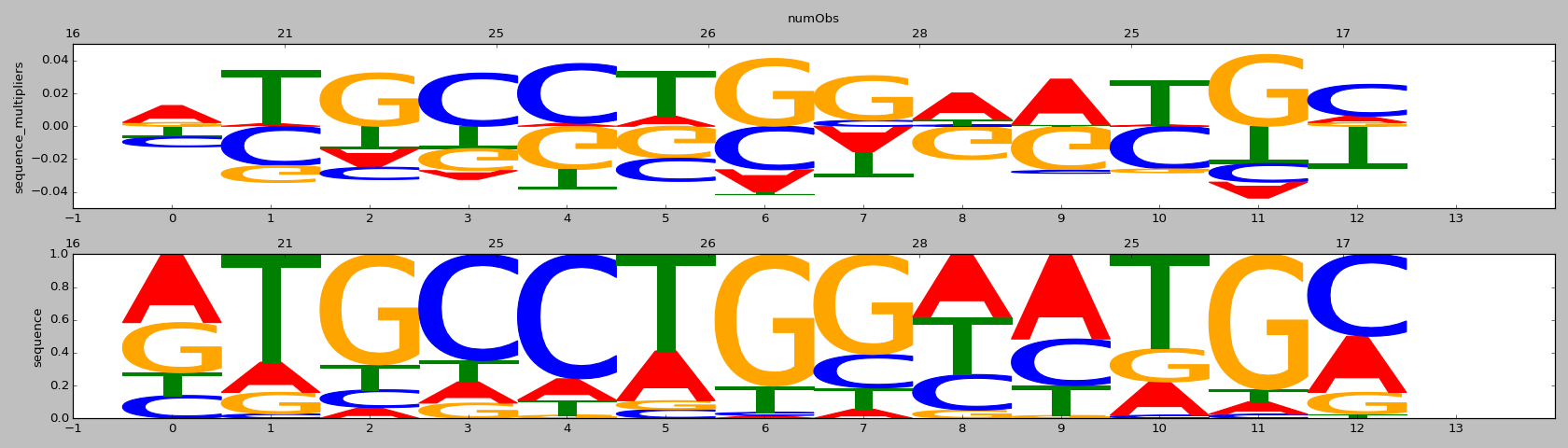
| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.75 |
TEAD2(TEA)/Py2T-Tead2-ChIP-Seq(GSE55709)/Homer |
 |
0.8 |
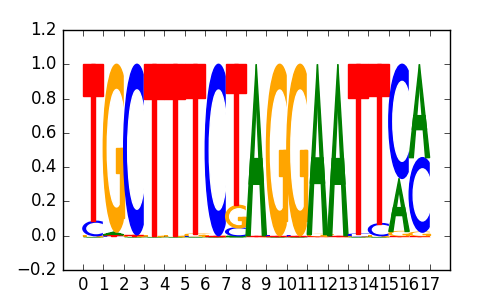
>BCL6B_2BCL6B_jolma_DBD_M1
|
 |
0.8 |
BCL6B |
 |
| 0.71 |
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer |
 |
0.79 |
>TCF12_disc6 TCF12_H1-hESC_encode-Myers_seq_hsa_r1:Trawler#1#Intergenic
|
 |
0.79 |
SOX10 |
 |
| 0.69 |
Zfp809(Zf)/ES-Zfp809-ChIP-Seq(GSE70799)/Homer |
 |
0.79 |
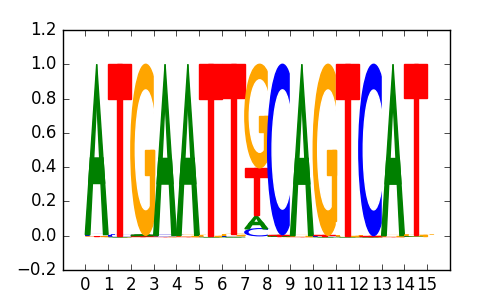
>SOX10_6SOX10_jolma_full_M758
|
 |
0.78 |
M2843_1.02.txt |
 |
| 0.69 |
STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer |
 |
0.78 |
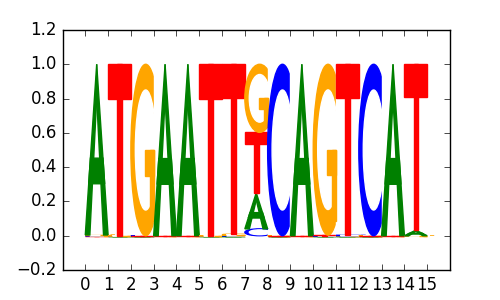
>SOX10_10Sox10_jolma_DBD_M803
|
 |
0.78 |
M6085_1.02.txt |
 |
| 0.69 |
Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer |
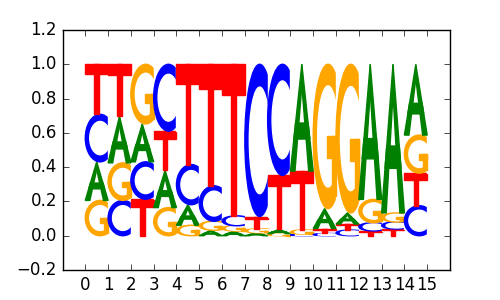 |
0.77 |
>SOX9_9SOX9_jolma_full_M796
|
 |
0.77 |
SOX9 |
 |
| 0.68 |
TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer |
 |
0.76 |
>TEAD1_4TEAD1_jolma_full_M287
|
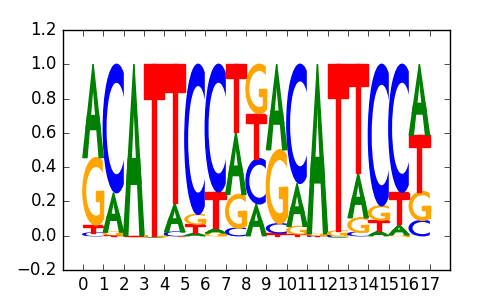 |
0.76 |
TEAD1 |
 |
| 0.68 |
Oct4:Sox17(POU,Homeobox,HMG)/F9-Sox17-ChIP-Seq(GSE44553)/Homer |
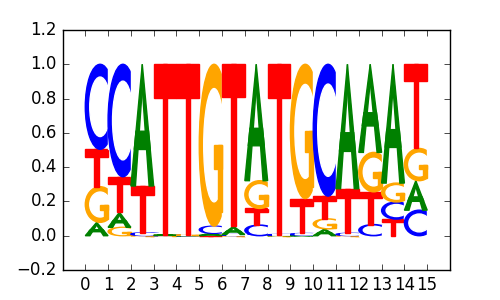 |
0.75 |
>TEAD3_1TEAD3_jolma_DBD_M288
|
 |
0.75 |
TEAD3 |
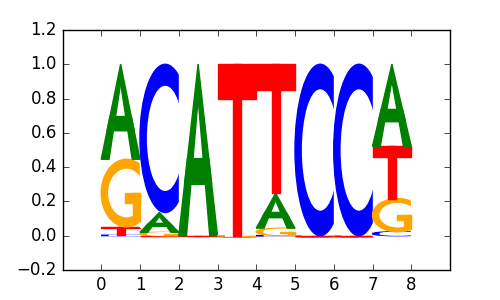 |
| 0.67 |
REST-NRSF(Zf)/Jurkat-NRSF-ChIP-Seq/Homer |
 |
0.74 |
>SOX18_4SOX18_jolma_full_M768
|
 |
0.74 |
SOX18 |
 |
| 0.67 |
Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer |
 |
0.73 |
>SOX7_4SOX7_jolma_full_M782
|
 |
0.73 |
SOX7 |
 |
| 0.67 |
Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer |
 |
0.72 |
>STAT_known16STAT3_3Stat3_jaspar_MA0144.1
|
 |
0.72 |
M2265_1.02.txt |
 |