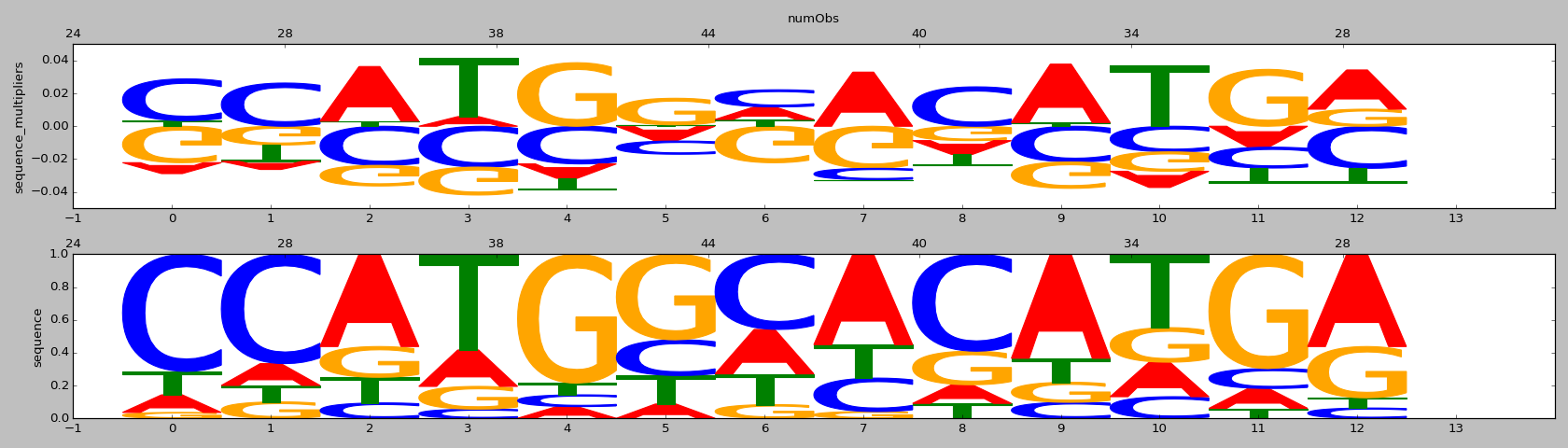
| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.68 |
Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer |
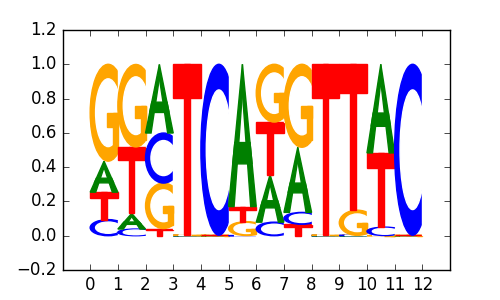 |
0.73 |
>NFATC1_3NFATC1_jolma_full_M188
|
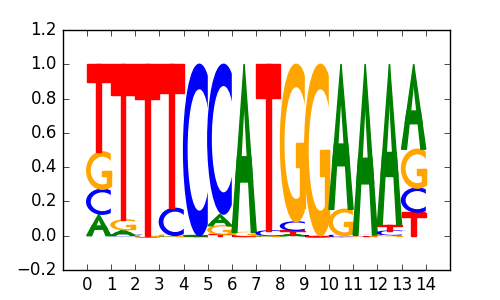 |
0.73 |
NFATC1 |
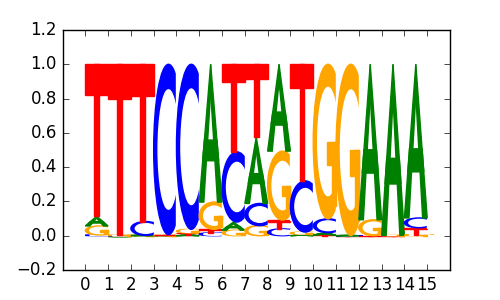 |
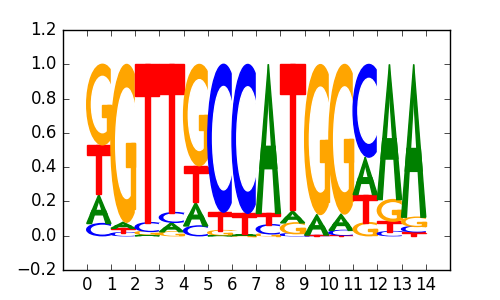
| 0.67 |
X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer |
 |
0.71 |
>BACH1_1Bach1_transfac_M00495
|
 |
0.72 |
M0621_1.02.txt |
 |
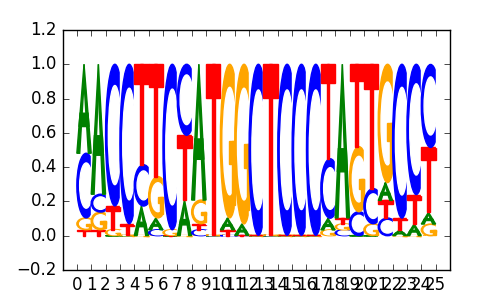
| 0.66 |
ZNF16(Zf)/HEK293-ZNF16.GFP-ChIP-Seq(GSE58341)/Homer |
 |
0.71 |
>NFE2_known2NFE2_2NFE2_jolma_DBD_M391
|
 |
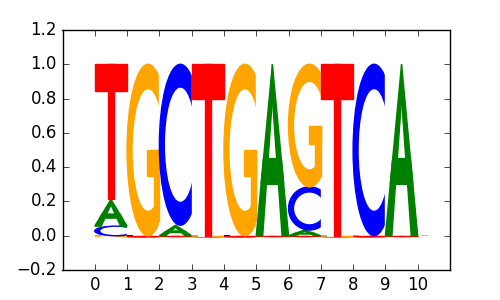
0.71 |
BACH1 |
 |
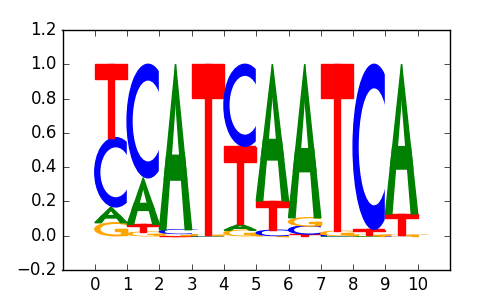
| 0.66 |
Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer |
 |
0.7 |
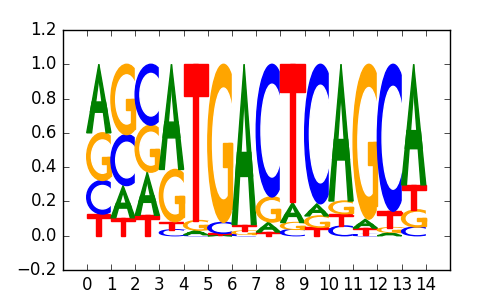
>JDP2_6Jdp2_jolma_DBD_M380
|
 |
0.71 |
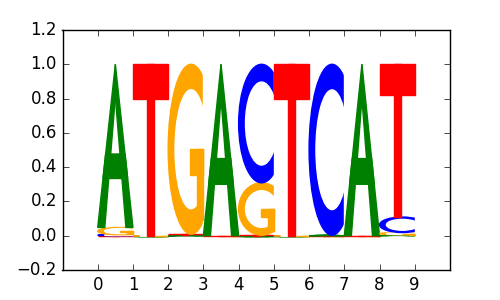
NFE2 |
 |
| 0.66 |
Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer |
 |
0.7 |
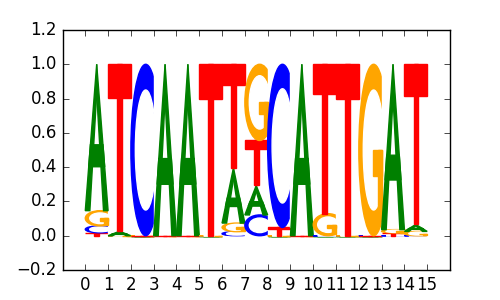
>SOX18_3SOX18_jolma_full_M767
|
 |
0.7 |
M6038_1.02.txt |
 |
| 0.66 |
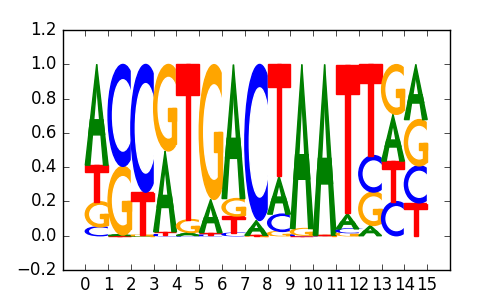
PAX3:FKHR-fusion(Paired,Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer |
 |
0.69 |
>THAP1_disc2 THAP1_K562_encode-Myers_seq_hsa_v041610.1-SC-98174_r1:Trawler#2#Intergenic
|
 |
0.7 |
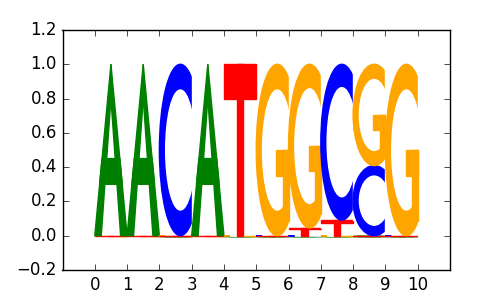
SOX18 |
 |
| 0.66 |
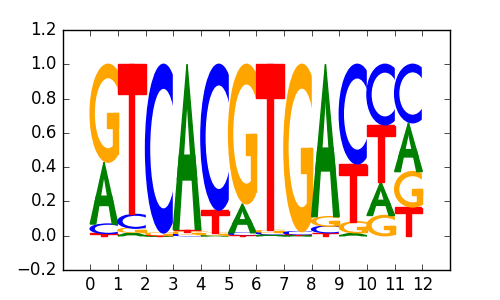
TFE3(bHLH)/MEF-TFE3-ChIP-Seq(GSE75757)/Homer |
 |
0.69 |
>TOPORS_1LUN-1_transfac_M00480
|
 |
0.7 |
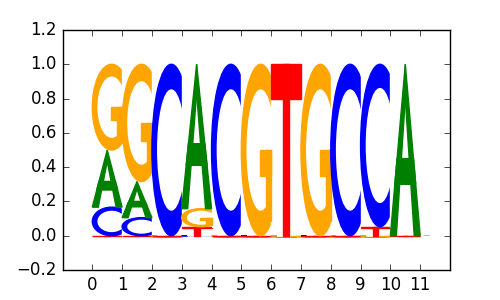
M5021_1.02.txt |
 |
| 0.65 |
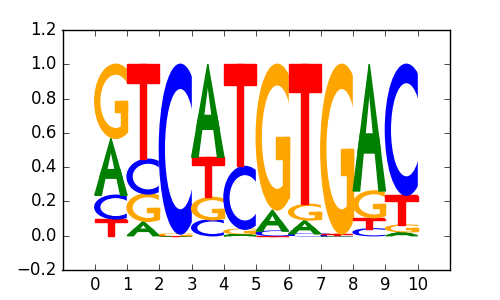
MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer |
 |
0.69 |
>ARNTL_1ARNTL_jolma_DBD_M306
|
 |
0.69 |
USF2 |
 |
| 0.65 |
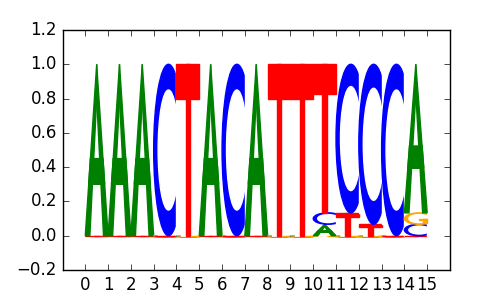
HOXA2(Homeobox)/mES-Hoxa2-ChIP-Seq(Donaldson_et_al.)/Homer |
 |
0.69 |
>SIX5_disc3 SIX5_H1-hESC_encode-Myers_seq_hsa_r1:Trawler#2#Intergenic
|
 |
0.69 |
TOPORS |
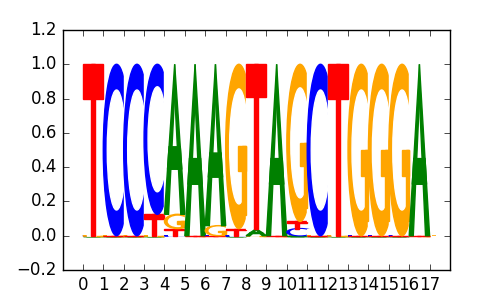 |
| 0.65 |
E-box(bHLH)/Promoter/Homer |
 |
0.69 |
>JDP2_4JDP2_jolma_full_M378
|
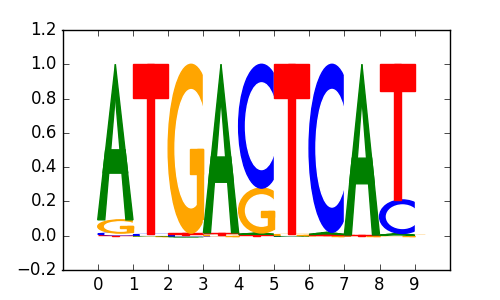 |
0.69 |
ARNTL |
 |