| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.84 |
Sp1(Zf)/Promoter/Homer |
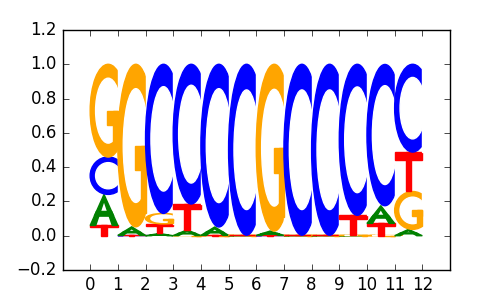 |
0.85 |
>EGR1_disc1 EGR1_H1-hESC_encode-Myers_seq_hsa_v041610.1_r1:MDscan#2#Intergenic
|
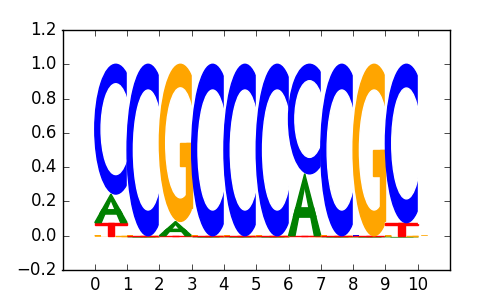 |
0.84 |
EGR1 |
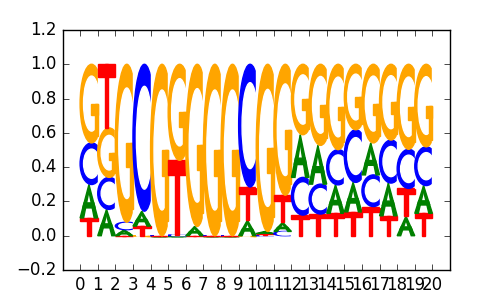 |
| 0.78 |
Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer |
 |
0.82 |
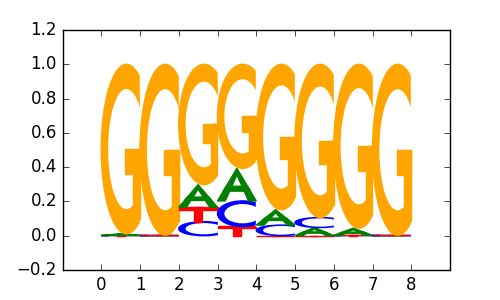
>EGR1_known7EGR1_4Egr1_bulyk_sc09-primary
|
 |
0.82 |
PATZ1 |
 |
| 0.77 |
Sp5(Zf)/mES-Sp5.Flag-ChIP-Seq(GSE72989)/Homer |
 |
0.82 |
>PATZ1_1MAZR_transfac_M00491
|
 |
0.82 |
EGR1 |
 |
| 0.77 |
Klf9(Zf)/GBM-Klf9-ChIP-Seq(GSE62211)/Homer |
 |
0.81 |
>HF1H3B_1UF1H3BETA_transfac_M01068
|
 |
0.81 |
SP4 |
 |
| 0.75 |
KLF6(Zf)/PDAC-KLF6-ChIP-Seq(GSE64557)/Homer |
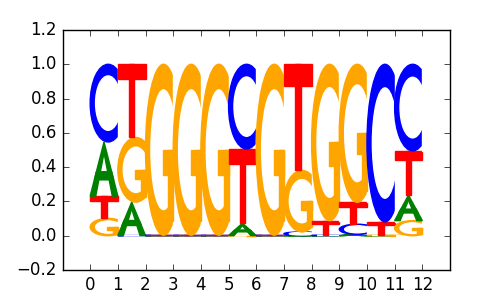 |
0.8 |
>ZNF219_1ZNF219_transfac_M01122
|
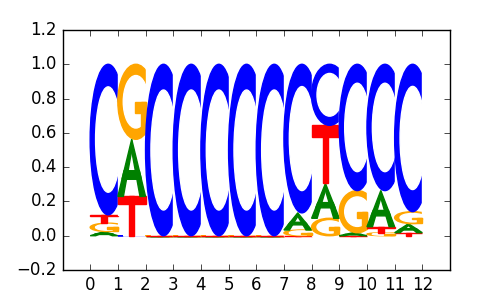 |
0.81 |
M4876_1.02.txt |
 |
| 0.75 |
KLF3(Zf)/MEF-Klf3-ChIP-Seq(GSE44748)/Homer |
 |
0.8 |
>SP2_disc3 SP2_K562_encode-Myers_seq_hsa_v041610.2-SC-643_r1:Trawler#3#Intergenic
|
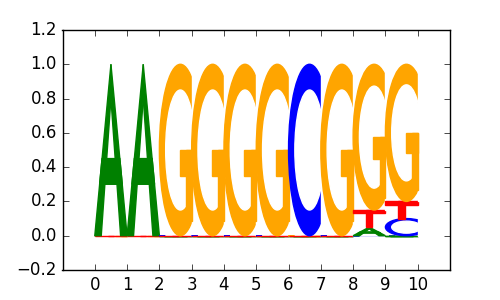 |
0.8 |
SP4 |
 |
| 0.74 |
KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer |
 |
0.8 |
>SP4_2SP4_jolma_full_M31
|
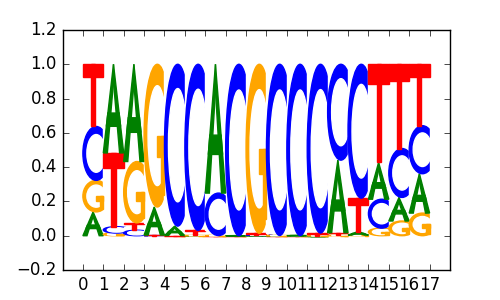 |
0.8 |
EGR1 |
 |
| 0.72 |
BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer |
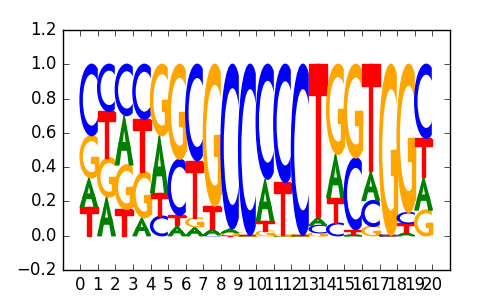 |
0.8 |
>ZNF740_3ZNF740_jolma_full_M51
|
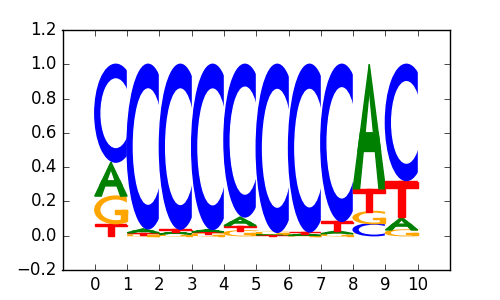 |
0.8 |
EGR1 |
 |
| 0.72 |
KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer |
 |
0.79 |
>TATA_disc4 TBP_K562_encode-Snyder_seq_hsa_IgG-mus_r1:MEME#1#Intergenic
|
 |
0.8 |
ZNF740 |
 |
| 0.71 |
Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer |
 |
0.79 |
>EGR1_known4EGR_2KROX_transfac_M00982
|
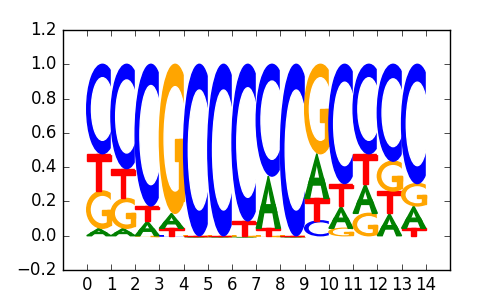 |
0.79 |
M4870_1.02.txt |
 |

































