![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | H7hESC-p7c7r7-trimmed-pair1.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 3660917 |
| Filtered Sequences | 0 |
| Sequence length | 44-151 |
| %GC | 46 |
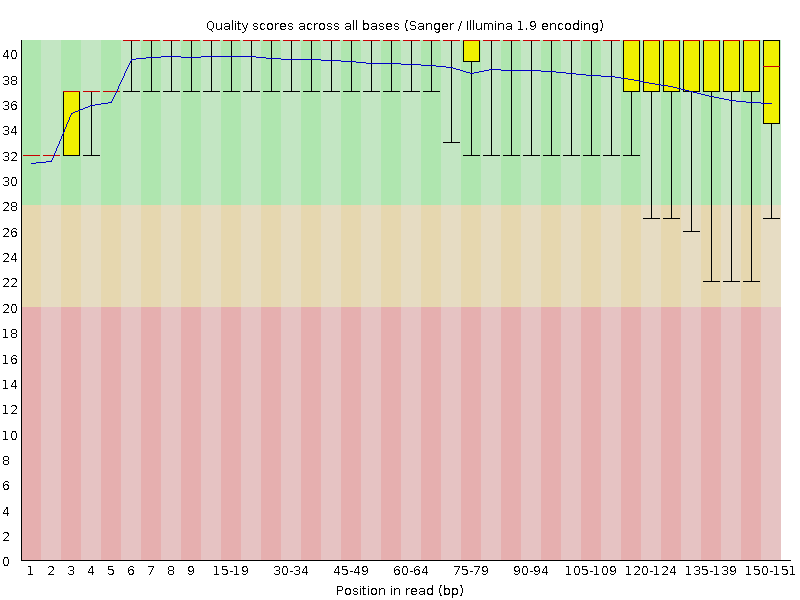
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
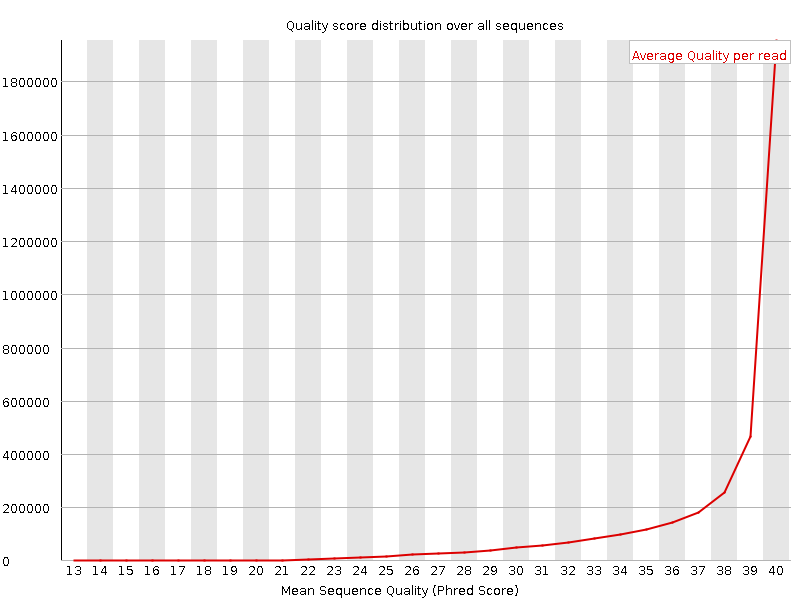
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
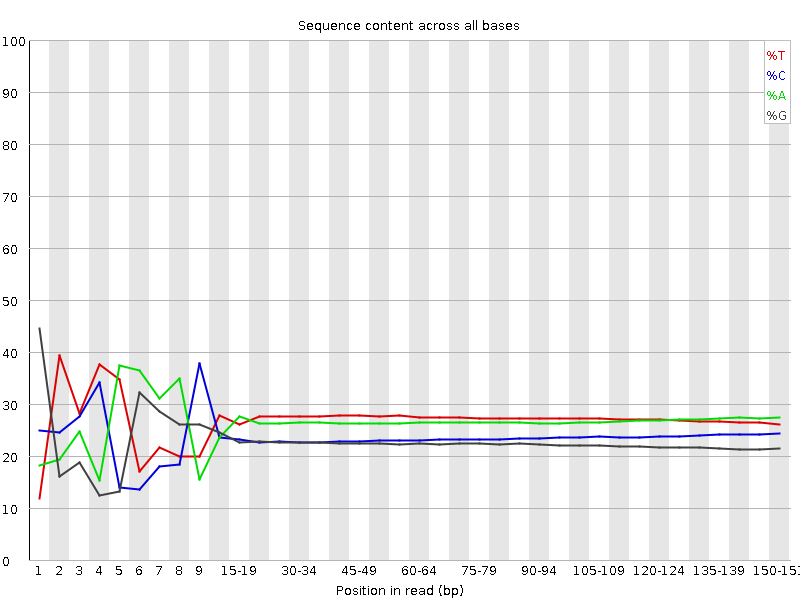
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
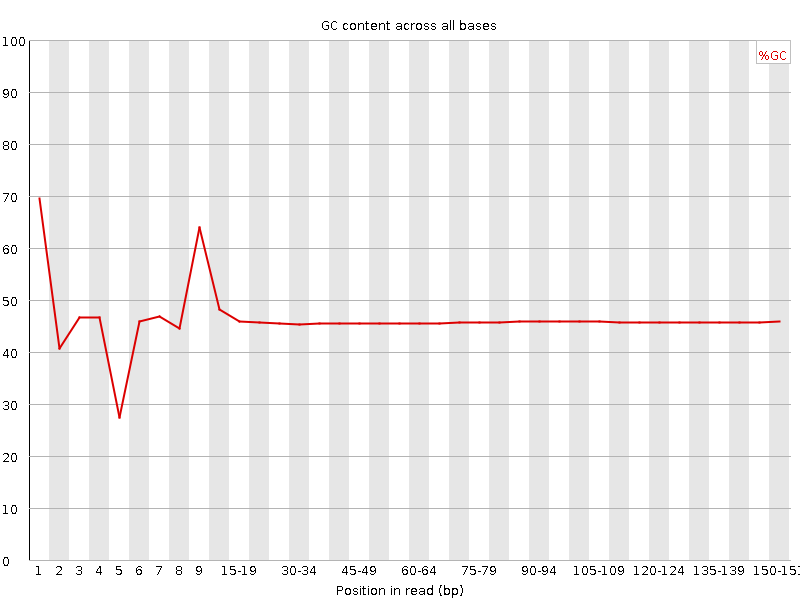
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[OK]](Icons/tick.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
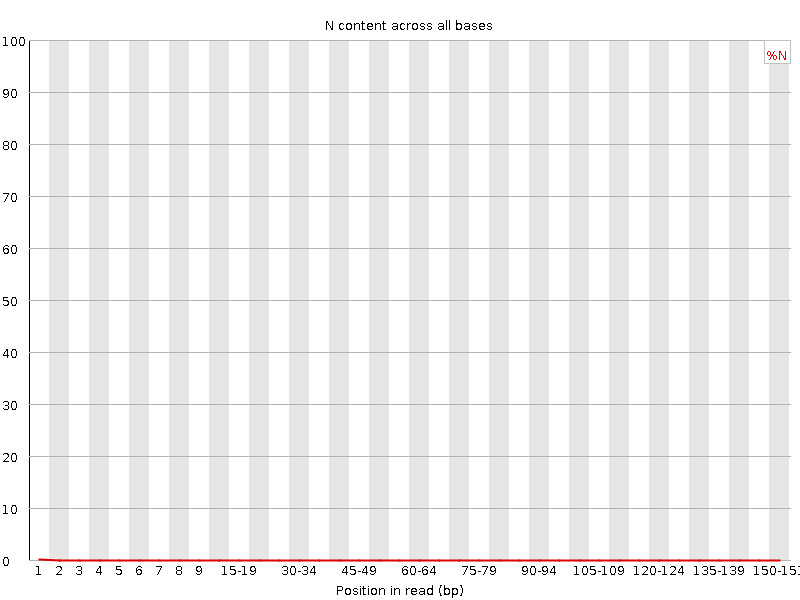
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
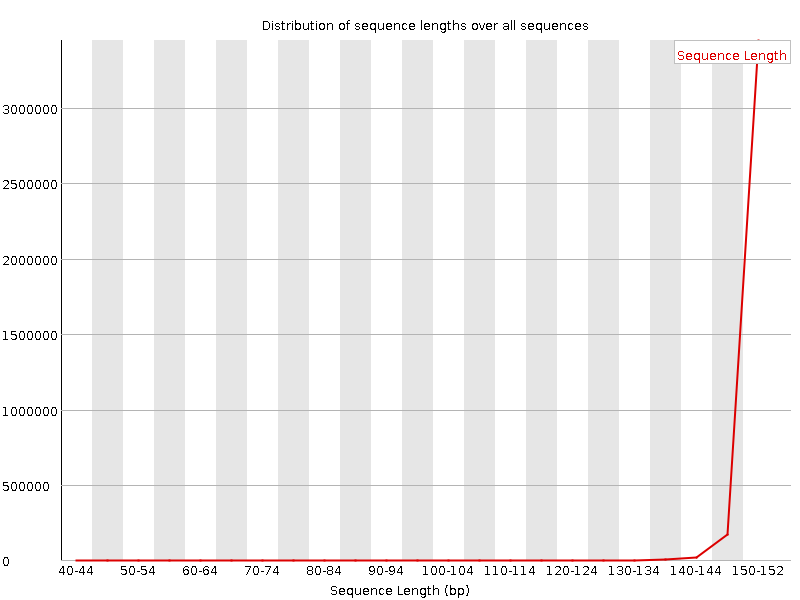
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GTATCAACGCAGAGTACTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | 11903 | 0.32513711728509553 | No Hit |
| TATCAACGCAGAGTACTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | 6054 | 0.16536840359942603 | No Hit |
| GGTATCAACGCAGAGTACTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | 5524 | 0.15089115650532364 | No Hit |
| CTTATACACATCTCCGAGCCCACGAGACAGGCAGAAATCTCGTATGCCGT | 3799 | 0.10377181454810365 | No Hit |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
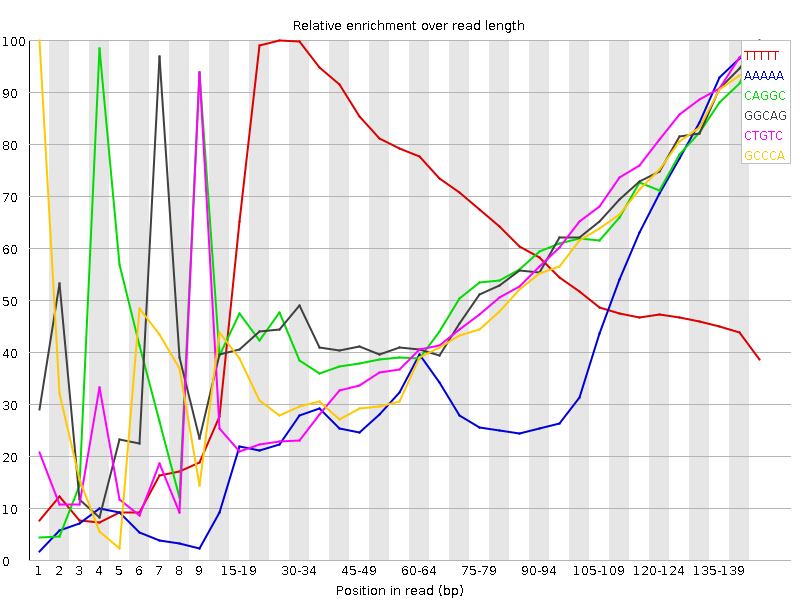
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| TTTTT | 5731960 | 7.055414 | 11.38118 | 25-29 |
| AAAAA | 3368495 | 4.664425 | 11.892793 | 145-147 |
| CAGGC | 1213220 | 3.0190477 | 5.46257 | 145-147 |
| GGCAG | 1138885 | 2.9665046 | 5.3221893 | 145-147 |
| CTGTC | 1359250 | 2.7240386 | 5.323158 | 145-147 |
| GCCCA | 1138635 | 2.70695 | 5.3052673 | 1 |
| GAGCC | 1081420 | 2.691069 | 5.797527 | 145-147 |
| GTCTC | 1293010 | 2.5912886 | 5.3124957 | 145-147 |
| GCAGA | 1155455 | 2.5407317 | 10.569439 | 9 |
| AGCCC | 1064800 | 2.5314174 | 5.383128 | 145-147 |
| ACAGG | 1114905 | 2.451566 | 5.382241 | 8 |
| ATCTC | 1448610 | 2.450789 | 5.918172 | 145-147 |
| ACACA | 1374835 | 2.43816 | 6.849892 | 6 |
| CTGGG | 940840 | 2.3936043 | 7.9563966 | 4 |
| GCTGG | 935105 | 2.3790138 | 5.2607284 | 3 |
| GGAGG | 871475 | 2.376046 | 5.1385775 | 8 |
| CAGAA | 1276640 | 2.3698165 | 5.0430737 | 4 |
| CCAGG | 934380 | 2.3251662 | 9.891681 | 3 |
| GGCTG | 907160 | 2.3079185 | 6.7867146 | 7 |
| CCTGG | 928775 | 2.25742 | 8.447738 | 3 |
| CCCAG | 942700 | 2.2411413 | 7.793834 | 2 |
| CTGCT | 1099110 | 2.2026985 | 5.4210224 | 9 |
| AGGAG | 951625 | 2.1903138 | 6.676524 | 5 |
| GACAG | 983890 | 2.163477 | 6.581518 | 7 |
| TGCTG | 1029810 | 2.1602583 | 5.8055673 | 2 |
| CAGCA | 999005 | 2.0986433 | 6.155414 | 4 |
| GCCTG | 853870 | 2.0753608 | 5.003109 | 7 |
| AGAAG | 1036750 | 2.0144434 | 5.1590934 | 5 |
| TCCTG | 1001835 | 2.0077522 | 8.551227 | 2 |
| AGACA | 1076360 | 1.9980388 | 5.0131407 | 6 |
| CAGGA | 899980 | 1.9789672 | 8.800352 | 4 |
| AGCAG | 895030 | 1.9680825 | 5.11466 | 7 |
| CAGAG | 894240 | 1.9663454 | 10.6513605 | 4 |
| CTCCT | 1019925 | 1.9527534 | 6.3004446 | 1 |
| TGGGG | 714605 | 1.9029938 | 10.002864 | 6 |
| TGGAG | 844935 | 1.899482 | 6.3629074 | 5 |
| GGCCA | 739845 | 1.8410738 | 5.3317156 | 1 |
| CTGGA | 853630 | 1.8333561 | 7.4663424 | 4 |
| GGGAG | 672300 | 1.8330023 | 5.6222553 | 7 |
| TACAC | 1042245 | 1.8053143 | 6.72981 | 5 |
| GGAGA | 780975 | 1.797536 | 5.0432477 | 6 |
| CAGGG | 681110 | 1.7741176 | 8.329368 | 4 |
| TGCAG | 821435 | 1.7642103 | 6.1046977 | 2 |
| CCCTG | 759390 | 1.7633237 | 5.846923 | 2 |
| TCCAG | 851990 | 1.748143 | 7.7686844 | 2 |
| CACAG | 825220 | 1.7335674 | 6.643426 | 7 |
| CTCTG | 847990 | 1.6994354 | 5.099261 | 2 |
| CTGTG | 805910 | 1.6905776 | 8.236261 | 9 |
| CAGTG | 784560 | 1.6850133 | 6.8565593 | 9 |
| AGAGG | 724705 | 1.6680217 | 5.088804 | 5 |
| CACTG | 810345 | 1.6626943 | 6.7324553 | 7 |
| CTCCA | 846555 | 1.6594452 | 5.6893263 | 1 |
| CATGG | 768760 | 1.6510793 | 14.513777 | 4 |
| TGGGC | 634795 | 1.6149908 | 5.3040805 | 5 |
| AGGTG | 698120 | 1.5694302 | 5.2620115 | 7 |
| CCAGA | 732250 | 1.5382624 | 5.2681403 | 3 |
| TCAGG | 697960 | 1.4990209 | 5.502503 | 3 |
| CTTGG | 702670 | 1.4740086 | 5.2082973 | 3 |
| GGGCT | 573205 | 1.4582989 | 5.3059454 | 6 |
| CCTGA | 709665 | 1.4561156 | 5.1018825 | 3 |
| ACAGA | 764495 | 1.4191259 | 5.622734 | 8 |
| GGGTG | 530725 | 1.4133211 | 5.2219896 | 7 |
| AGGGC | 532315 | 1.3865446 | 5.1001506 | 5 |
| GGGGG | 428100 | 1.3826253 | 6.039635 | 7 |
| ACAGC | 657440 | 1.3811063 | 5.8164625 | 8 |
| ATGGG | 614215 | 1.3808051 | 13.80031 | 5 |
| GTCTT | 789085 | 1.3648484 | 5.844504 | 1 |
| GTGTG | 614105 | 1.3484223 | 6.122561 | 1 |
| TCTTG | 761295 | 1.3167812 | 5.2045593 | 2 |
| AACAG | 702880 | 1.3047507 | 5.415897 | 7 |
| ACTGG | 605745 | 1.3009691 | 5.245938 | 8 |
| CACAC | 634495 | 1.273399 | 5.086038 | 5 |
| ACATG | 678915 | 1.2309294 | 11.955907 | 3 |
| ACTGC | 593625 | 1.2180207 | 5.032796 | 8 |
| ACTGT | 685685 | 1.214266 | 6.6401176 | 8 |
| GTGCT | 568730 | 1.1930392 | 5.7952237 | 1 |
| AGCAC | 559250 | 1.1748353 | 5.4623075 | 5 |
| TACAG | 632535 | 1.1468385 | 5.3278413 | 2 |
| GGACA | 514410 | 1.1311368 | 5.6933823 | 6 |
| AGGAC | 507490 | 1.1159204 | 6.202757 | 5 |
| AGAGT | 577100 | 1.0952255 | 5.97808 | 5 |
| GACTG | 501905 | 1.0779502 | 5.73558 | 7 |
| ACAGT | 590015 | 1.0697463 | 6.4395046 | 8 |
| ATCAA | 685570 | 1.0493252 | 7.7070036 | 3 |
| GTCCT | 518880 | 1.0398742 | 6.1909766 | 1 |
| GGACT | 475145 | 1.0204772 | 5.1389127 | 6 |
| GTGCA | 474150 | 1.0183402 | 6.219259 | 1 |
| GTCCA | 496035 | 1.0177821 | 6.6358256 | 1 |
| TGGAC | 471620 | 1.0129066 | 5.611239 | 5 |
| TCAAC | 559855 | 0.9697473 | 8.3434 | 4 |
| GAGTA | 501845 | 0.9524059 | 10.26252 | 1 |
| GTACA | 495210 | 0.8978569 | 16.070776 | 1 |
| TACTG | 503110 | 0.8909475 | 5.3705115 | 7 |
| AGTAC | 474650 | 0.86057997 | 7.4701557 | 2 |
| TACAT | 560635 | 0.8381272 | 10.323161 | 2 |
| CGCAG | 321905 | 0.8010473 | 11.187792 | 8 |
| TATCA | 511805 | 0.76512825 | 8.159565 | 2 |
| GTATT | 476425 | 0.7281661 | 5.0789995 | 1 |
| GTGTA | 376790 | 0.69843054 | 6.694242 | 1 |
| GGTAT | 330480 | 0.61258876 | 5.7957683 | 1 |
| GTATC | 312320 | 0.55308133 | 9.470641 | 1 |
| GTATA | 327895 | 0.51309675 | 6.0839787 | 1 |
| ACGCA | 226560 | 0.47594222 | 9.476969 | 7 |
| CAACG | 222990 | 0.4684426 | 9.447774 | 5 |
| AACGC | 190430 | 0.4000427 | 9.440074 | 6 |