Dup QC (all)
Flagstat QC (all, filtered)
PBC QC (all)
| Total Read Pairs | Distinct Read Pairs | One Read Pair | Two Read Pairs | NRF = Distinct/Total | PBC1 = OnePair/Distinct | PBC2 = OnePair/TwoPair | |
|---|---|---|---|---|---|---|---|
| rep1 | 20029755 | 18001272 | 16269123 | 1582725 | 0.898727 | 0.903776 | 10.279185 |
| rep2 | 18538368 | 16761658 | 15245573 | 1390304 | 0.904160 | 0.909550 | 10.965640 |
NRF (non redundant fraction)
PBC1 (PCR Bottleneck coefficient 1)
PBC2 (PCR Bottleneck coefficient 2)
PBC1 is the primary measure. Provisionally
- 0-0.5 is severe bottlenecking
- 0.5-0.8 is moderate bottlenecking
- 0.8-0.9 is mild bottlenecking
- 0.9-1.0 is no bottlenecking
Cross-correlation QC (all)
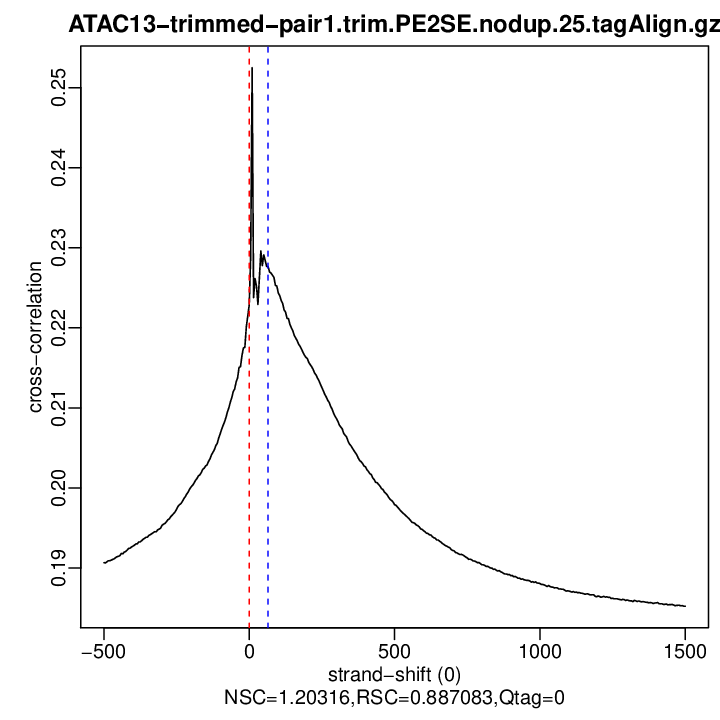
rep1 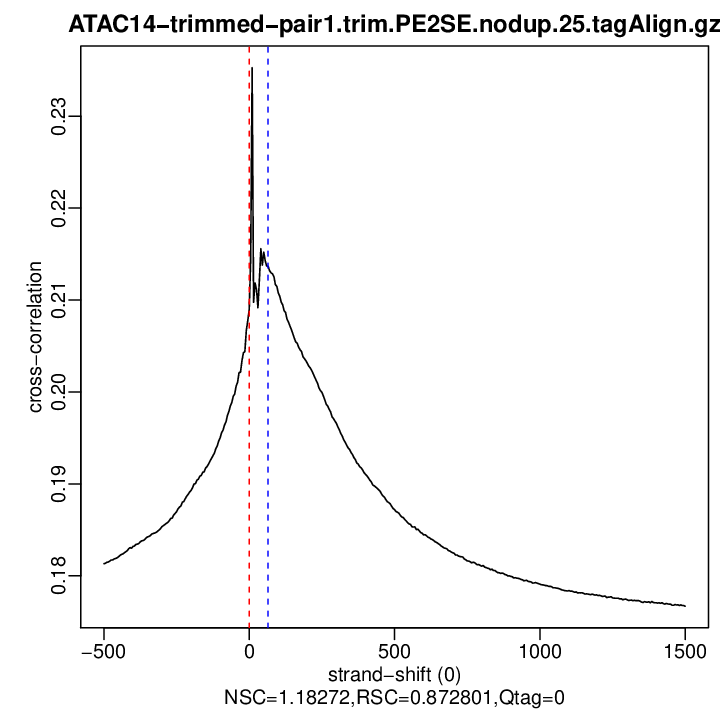
rep2
| numReads | estFragLen | corr_estFragLen | PhantomPeak | corr_phantomPeak | argmin_corr | min_corr | phantomPeakCoef | relPhantomPeakCoef | QualityTag | |
|---|---|---|---|---|---|---|---|---|---|---|
| rep1 | 18000883 | 0 | 0.222849954004333 | 65 | 0.2276399 | 1500 | 0.1852198 | 1.203165 | 0.8870829 | 0 |
| rep2 | 16761251 | 0 | 0.208967806844454 | 65 | 0.2136727 | 1500 | 0.1766845 | 1.182718 | 0.8728015 | 0 |
Normalized strand cross-correlation coefficient (NSC) = col9 in outFile
Relative strand cross-correlation coefficient (RSC) = col10 in outFile
Estimated fragment length = col3 in outFile, take the top value
Important columns highlighted, but all/whole file can be stored for display


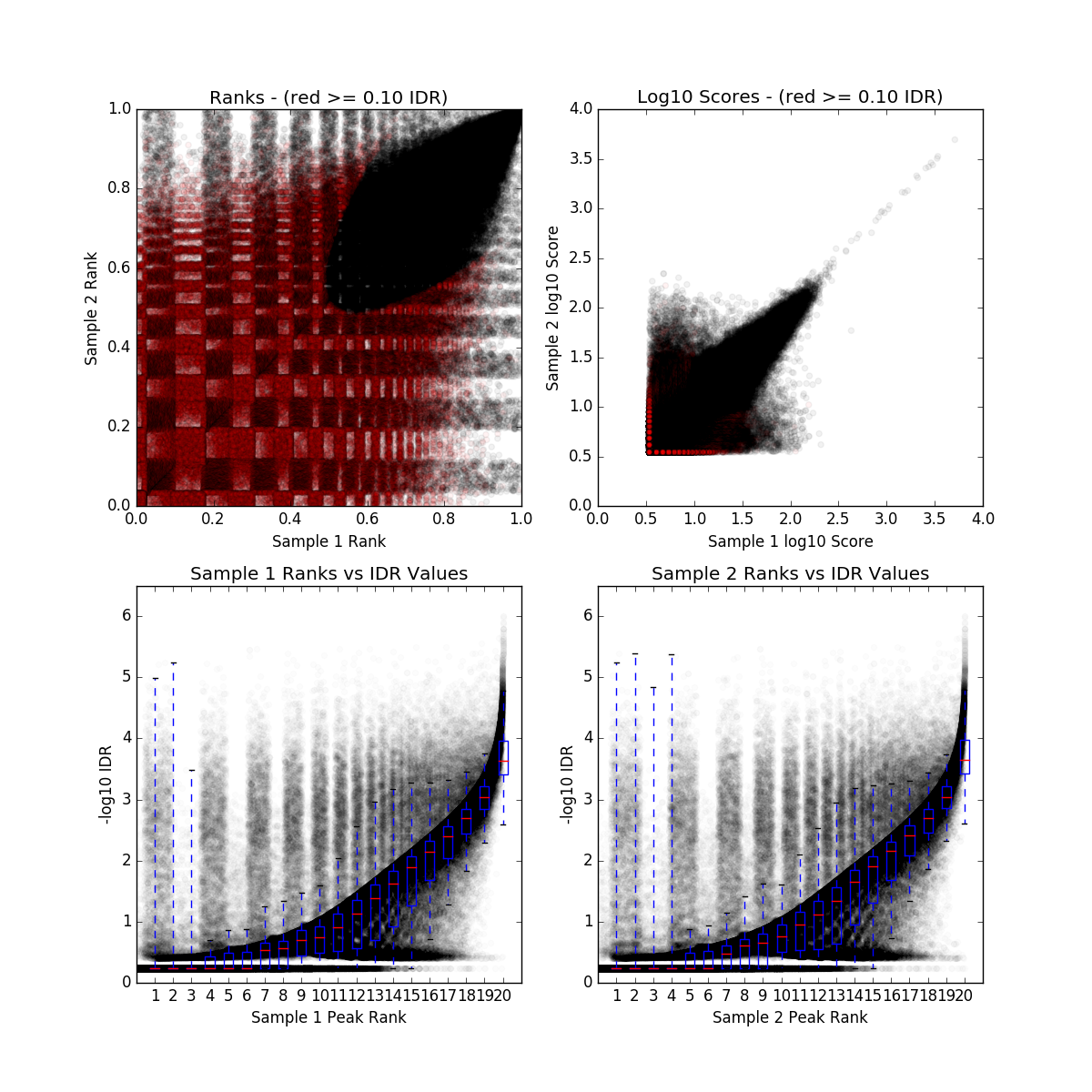
IDR QC (idr)
IDR_final.qc
| Nt | Np | conservative_set | optimal_set | rescue_ratio | self_consistency_ratio | reproducibility |
|---|---|---|---|---|---|---|
| 116312 | 0 | rep1-rep2 | rep1-rep2 | Infinity | NaN | 0 |