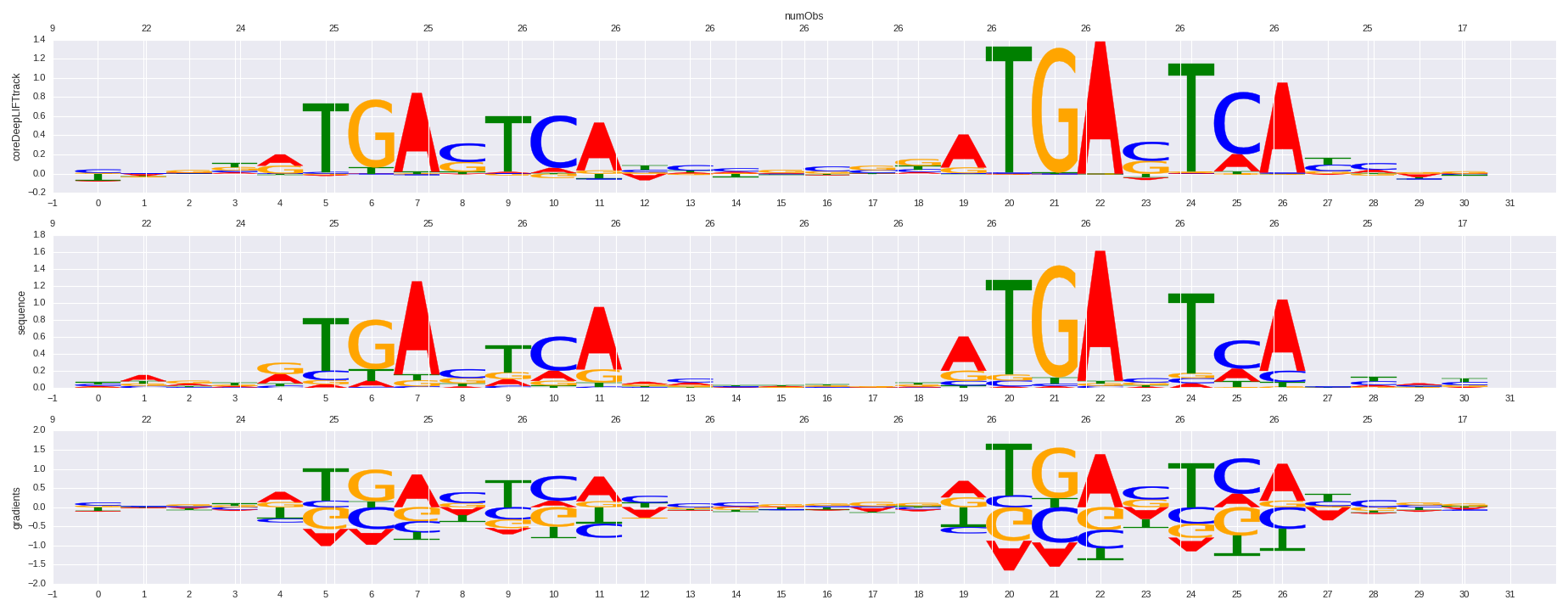
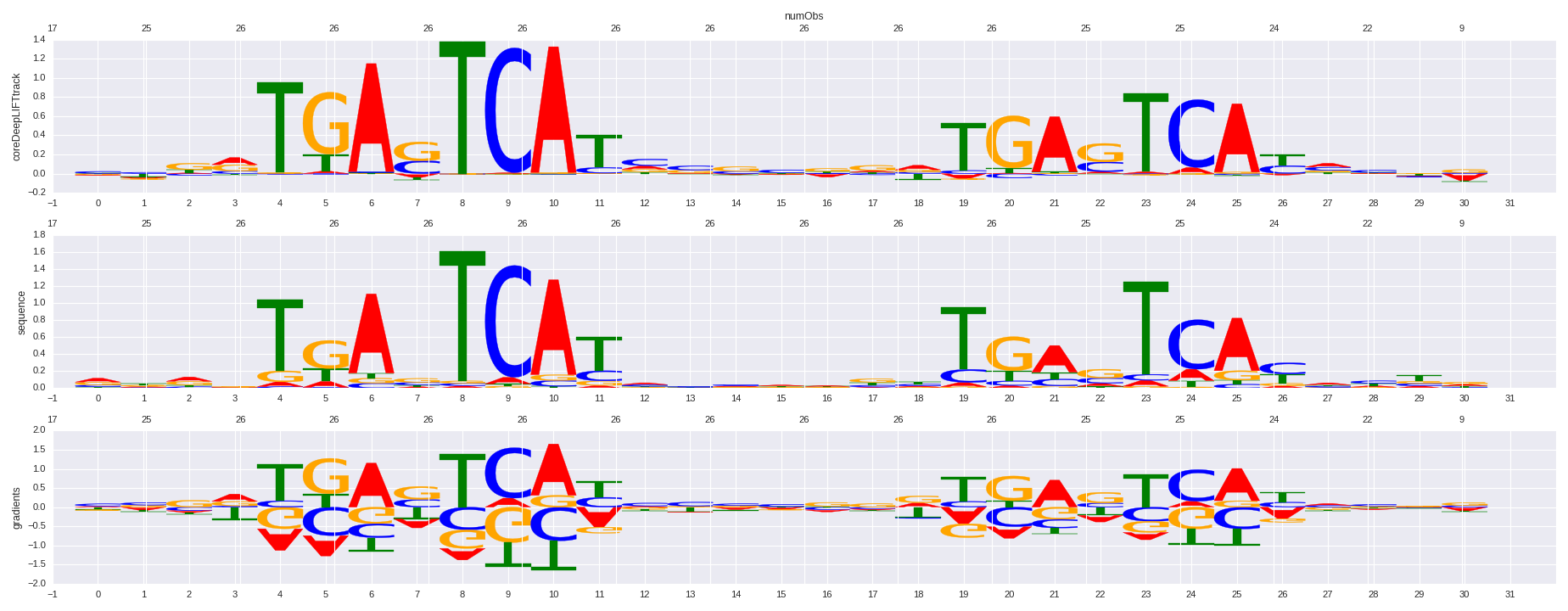
| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.704797735779 |
Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer |
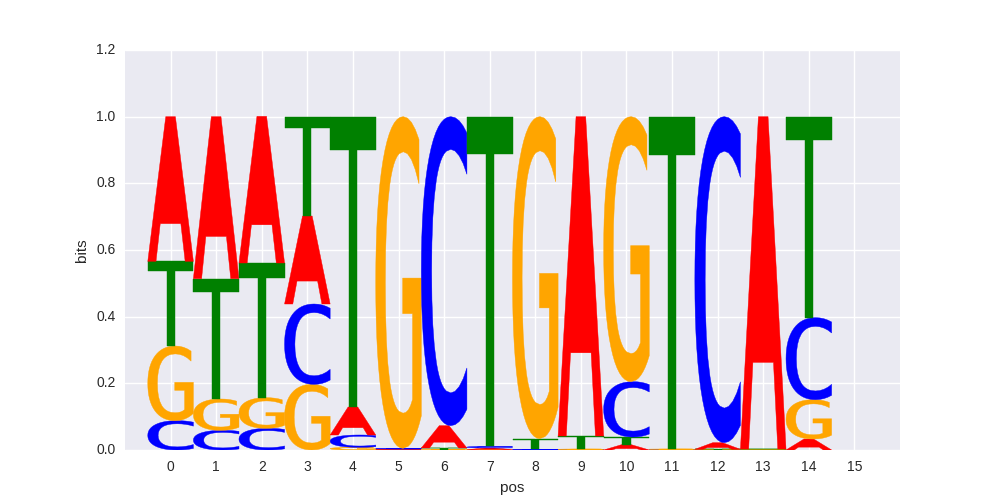 |
0.67462278954 |
>NFE2L1::MAFG_1TCF11:MafG_transfac_M00284
|
 |
0.737917981342 |
M2428_1.02.txt |
 |
| 0.621045638619 |
PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer |
 |
0.658278025085 |
>ATF3_known5CREB1_4Tax/CREB_transfac_M00115
|
 |
0.736666625767 |
JUND |
 |
| 0.618762037466 |
NFAT:AP1(RHD,bZIP)/Jurkat-NFATC1-ChIP-Seq(Jolma_et_al.)/Homer |
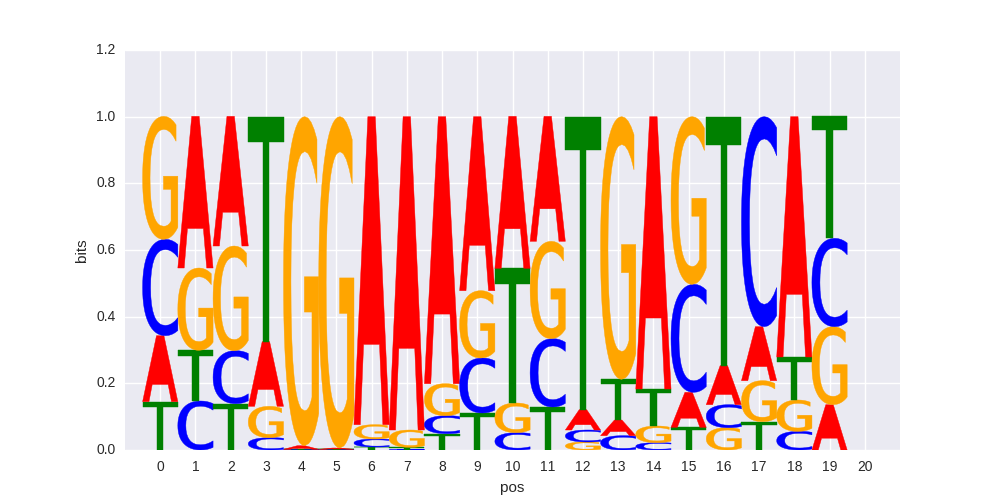 |
0.652217153172 |
>MAFG_1MAFG_jolma_full_M383
|
 |
0.735993750566 |
JUND |
 |
| 0.613224532761 |
OCT:OCT(POU,Homeobox,IR1)/NPC-Brn2-ChIP-Seq(GSE35496)/Homer |
 |
0.651322880682 |
>TBX1_4TBX1_jolma_DBD_M268
|
 |
0.734961706604 |
FOS |
 |
| 0.612120879288 |
Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer |
 |
0.647903214525 |
>T_1Brachyury_transfac_M00150
|
 |
0.732644583754 |
FOSL2 |
 |
| 0.601999187581 |
ZFP3(Zf)/HEK293-ZFP3.GFP-ChIP-Seq(GSE58341)/Homer |
 |
0.641252215281 |
>VENTX_2VENTX_jolma_DBD_M650
|
 |
0.731805130505 |
JUND |
 |
| 0.598506937555 |
Hoxc9(Homeobox)/Ainv15-Hoxc9-ChIP-Seq(GSE21812)/Homer |
 |
0.637707172656 |
>SOX15_4SOX15_jolma_full_M765
|
 |
0.728734042756 |
JUNB |
 |
| 0.595455257041 |
Pit1+1bp(Homeobox)/GCrat-Pit1-ChIP-Seq(GSE58009)/Homer |
 |
0.63408973718 |
>MAF_known11MAFB_4Mafb_jolma_DBD_M389
|
 |
0.726268433449 |
JUN |
 |
| 0.592464702353 |
ZNF16(Zf)/HEK293-ZNF16.GFP-ChIP-Seq(GSE58341)/Homer |
 |
0.631489052948 |
>PAX9_1PAX9_jolma_DBD_M204
|
 |
0.726005072834 |
SMARCC1 |
 |
| 0.590533613465 |
PAX5(Paired,Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer |
 |
0.629695125786 |
>SOX9_5SOX9_jolma_full_M792
|
 |
0.725954465148 |
JUN |
 |