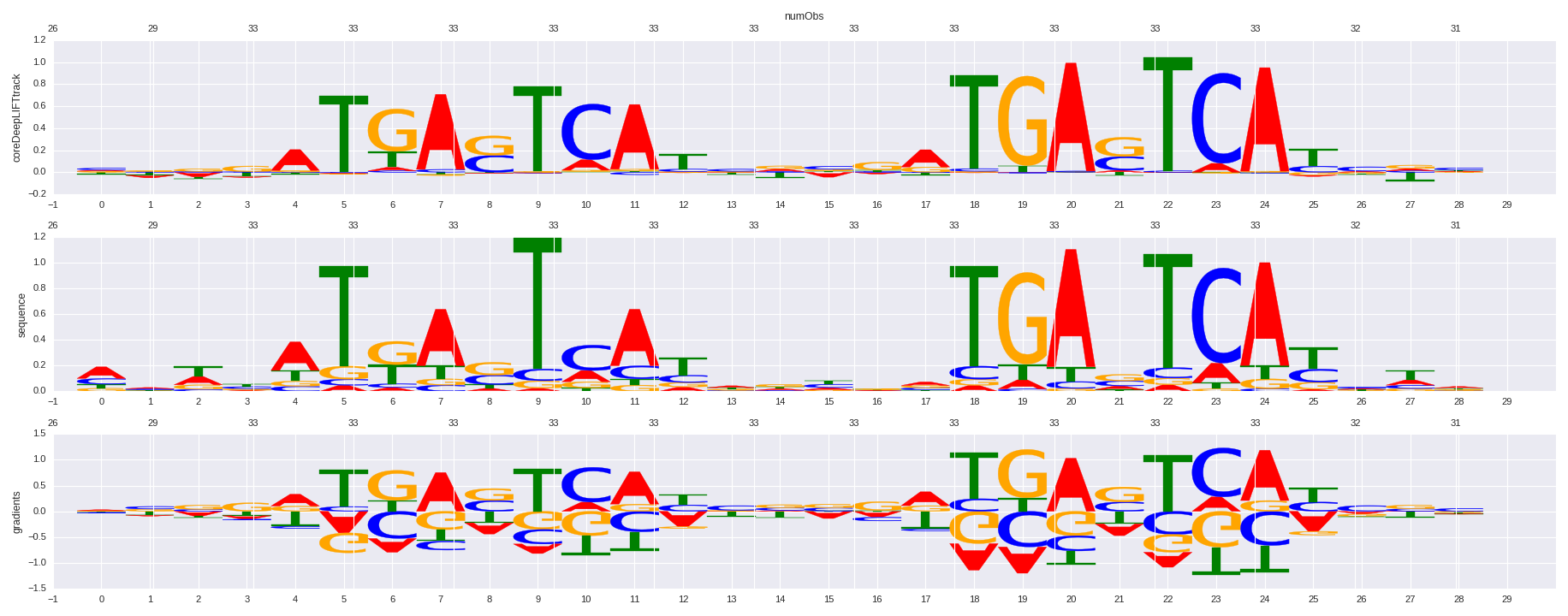
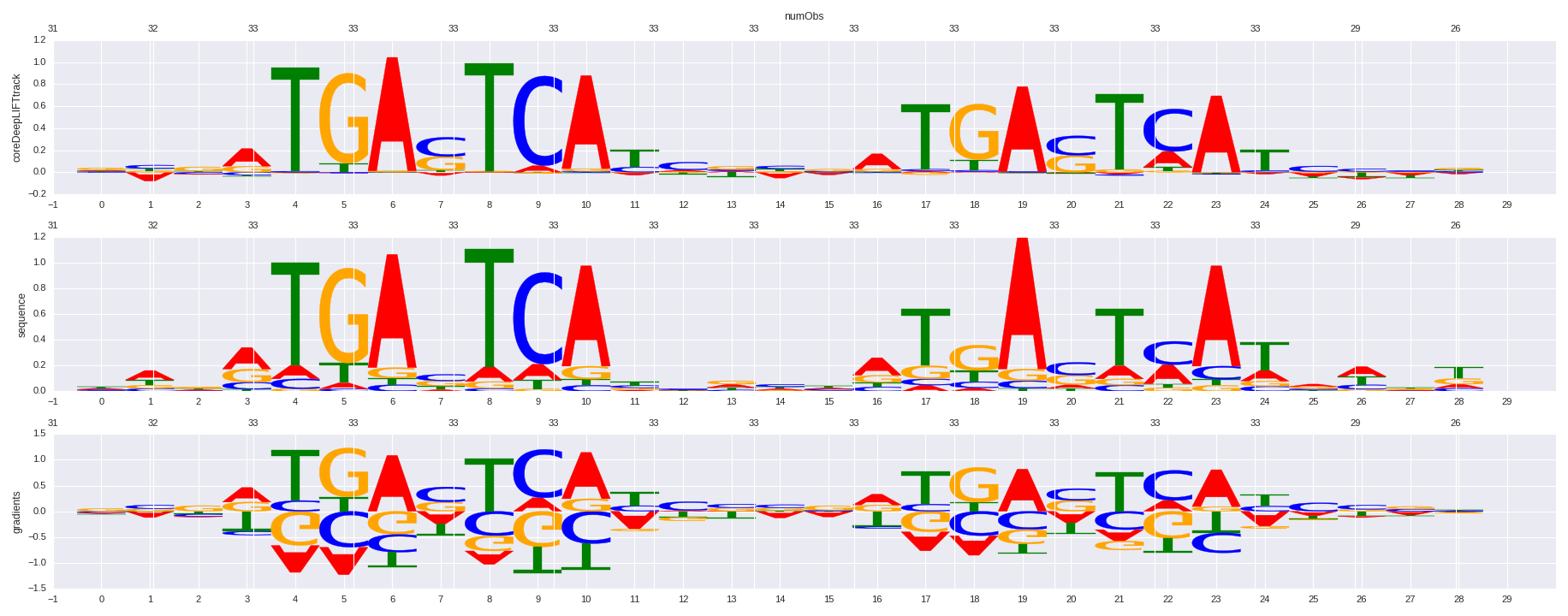
| HOMER correlation |
HOMER motif name |
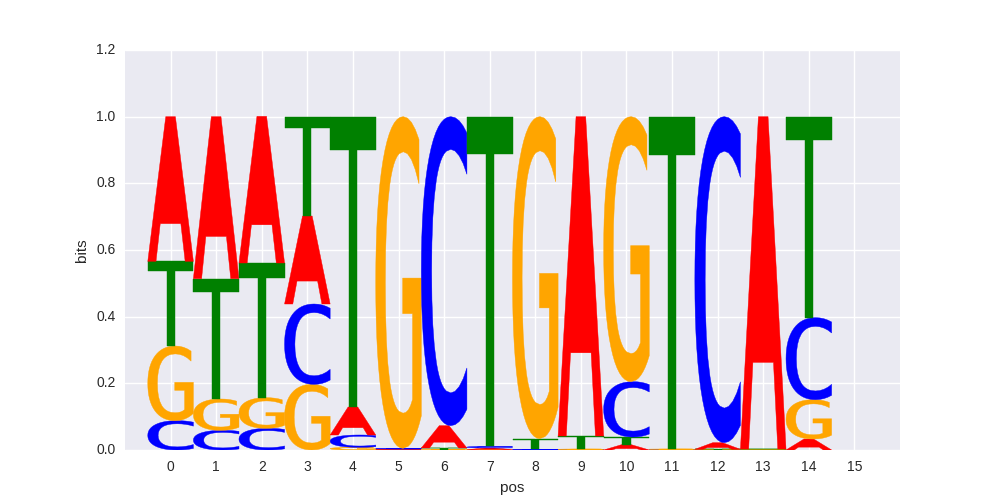
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
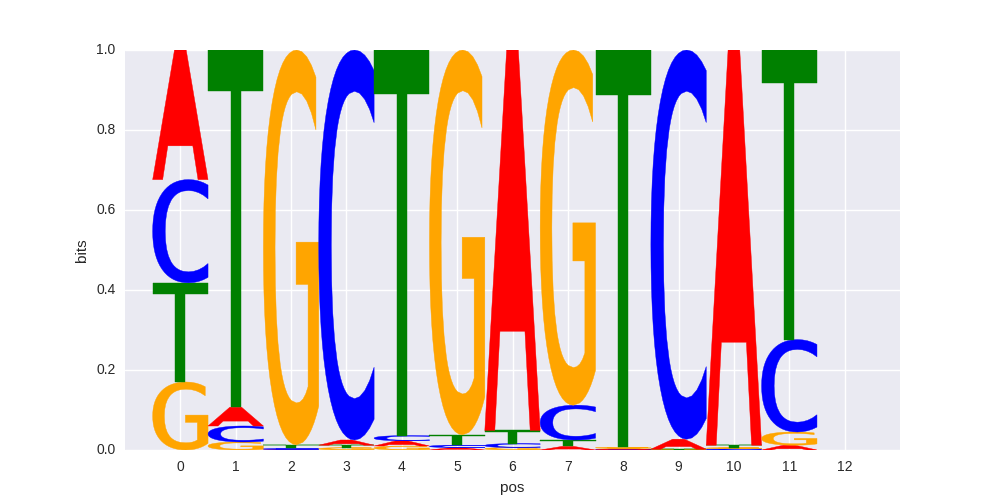
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.714925786478 |
Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer |
 |
0.728172138087 |
>BACH1_1Bach1_transfac_M00495
|
 |
0.728172140662 |
BACH1 |
 |
| 0.710764966853 |
Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer |
 |
0.708340488723 |
>BATF_disc1 BATF_GM12878_encode-Myers_seq_hsa_r1:AlignACE#1#Intergenic
|
 |
0.724272121764 |
JUND |
 |
| 0.631986416261 |
Fra2(bZIP)/Striatum-Fra2-ChIP-Seq(GSE43429)/Homer |
 |
0.680988891679 |
>SOX15_4SOX15_jolma_full_M765
|
 |
0.724055739181 |
JUND |
 |
| 0.628397723596 |
Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer |
 |
0.678284261371 |
>MAFF_1MAFF_jolma_DBD_M382
|
 |
0.72345973115 |
FOS |
 |
| 0.627356205558 |
Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer |
 |
0.666577245798 |
>ATF3_known5CREB1_4Tax/CREB_transfac_M00115
|
 |
0.721284730597 |
FOSL2 |
 |
| 0.627065321092 |
PAX3:FKHR-fusion(Paired,Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer |
 |
0.661106916387 |
>NFE2L2_2Nrf-2_transfac_M00821
|
 |
0.720337128398 |
JUND |
 |
| 0.624146999548 |
NFAT:AP1(RHD,bZIP)/Jurkat-NFATC1-ChIP-Seq(Jolma_et_al.)/Homer |
 |
0.654358310916 |
>MAFG_1MAFG_jolma_full_M383
|
 |
0.718748785295 |
JUNB |
 |
| 0.622657920603 |
NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer |
 |
0.641998673925 |
>NFE2L1::MAFG_1TCF11:MafG_transfac_M00284
|
 |
0.717263151287 |
JUN |
 |
| 0.621394983186 |
PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer |
 |
0.633485911218 |
>SOX10_10Sox10_jolma_DBD_M803
|
 |
0.717204979878 |
M2428_1.02.txt |
 |
| 0.61931442917 |
Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer |
 |
0.633275376528 |
>SOX10_6SOX10_jolma_full_M758
|
 |
0.715683115423 |
FOS |
 |