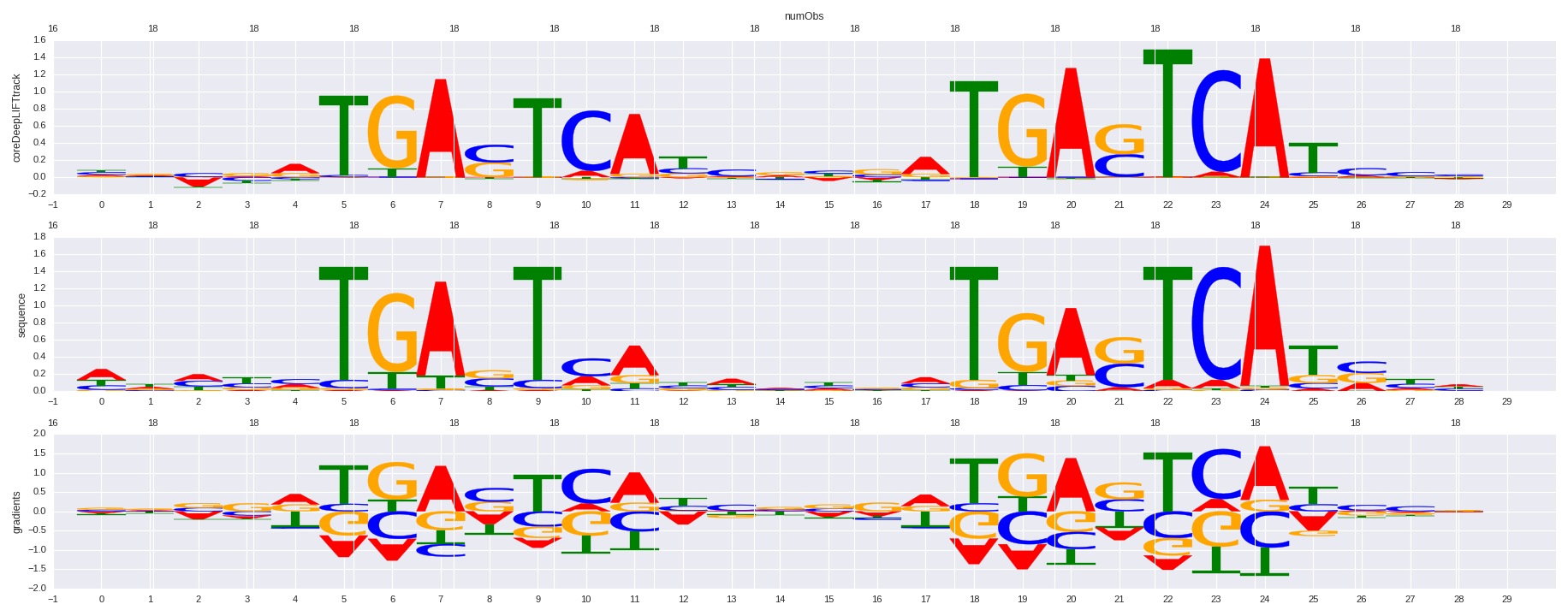
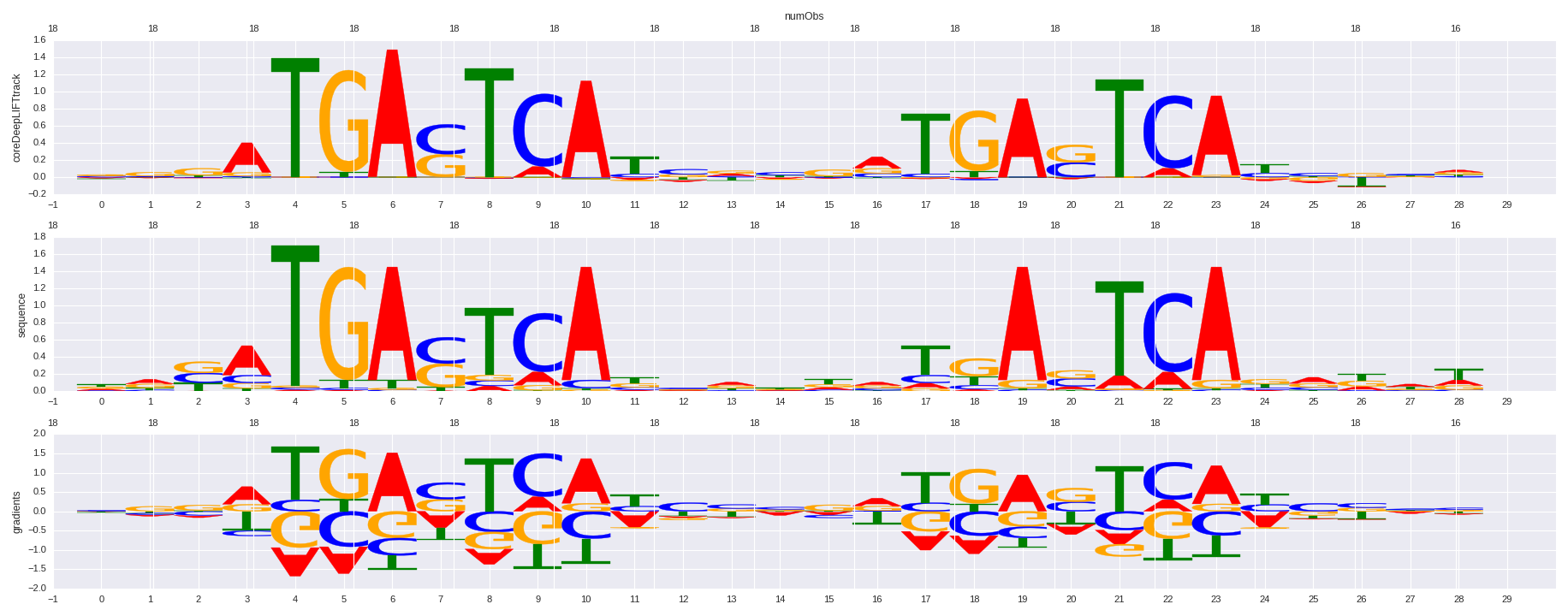
| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.711788860801 |
Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer |
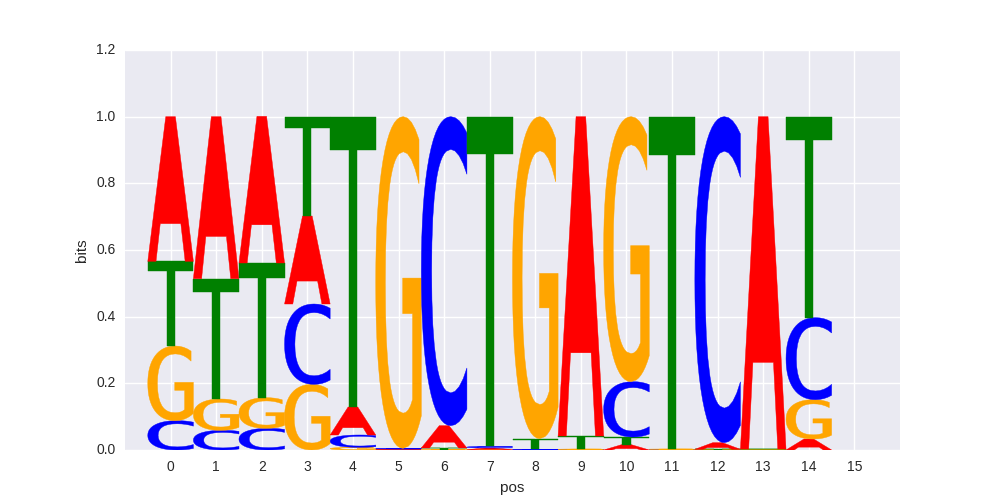 |
0.722840539898 |
>BACH1_1Bach1_transfac_M00495
|
 |
0.72284054201 |
BACH1 |
 |
| 0.707712644024 |
Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer |
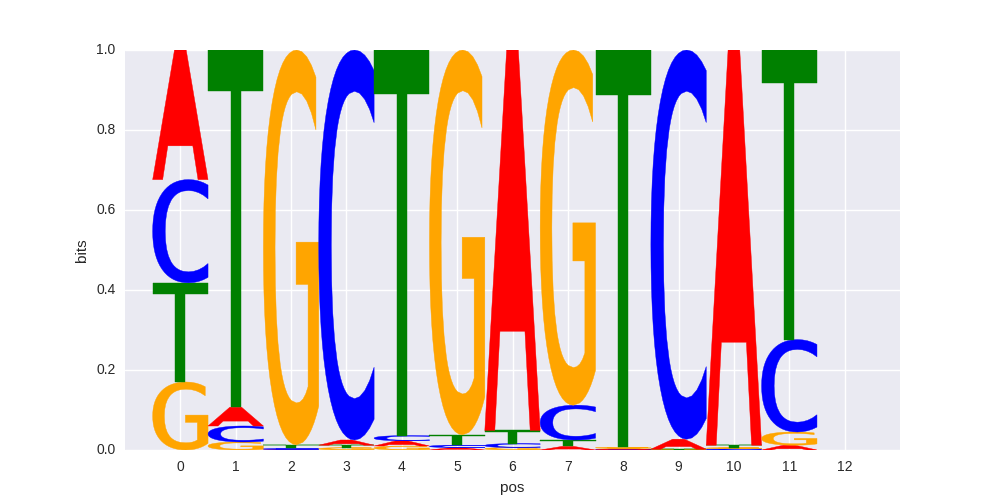 |
0.702629004095 |
>BATF_disc1 BATF_GM12878_encode-Myers_seq_hsa_r1:AlignACE#1#Intergenic
|
 |
0.720523703594 |
JUND |
 |
| 0.629540321381 |
Fra2(bZIP)/Striatum-Fra2-ChIP-Seq(GSE43429)/Homer |
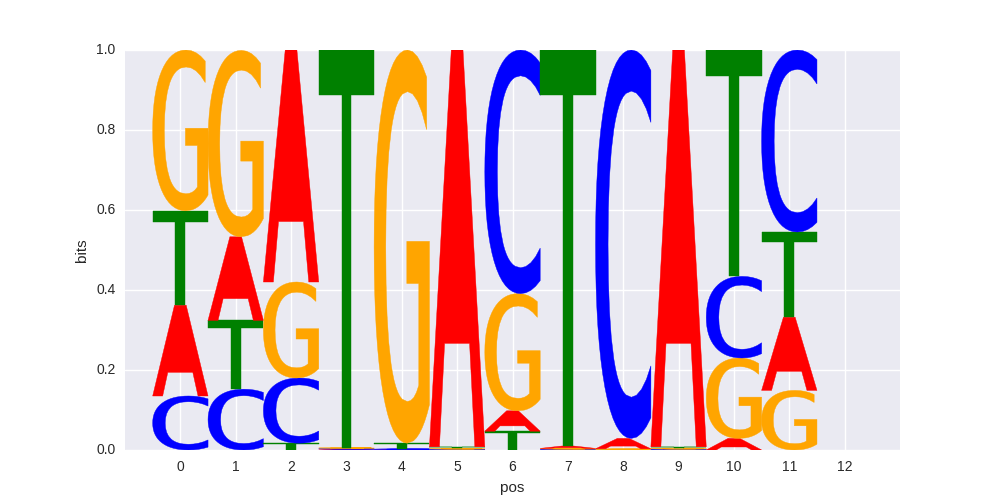 |
0.670767552658 |
>MAFF_1MAFF_jolma_DBD_M382
|
 |
0.717943560524 |
JUND |
 |
| 0.627804035041 |
Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer |
 |
0.669953456283 |
>SOX15_4SOX15_jolma_full_M765
|
 |
0.716477975612 |
FOS |
 |
| 0.626566693152 |
Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer |
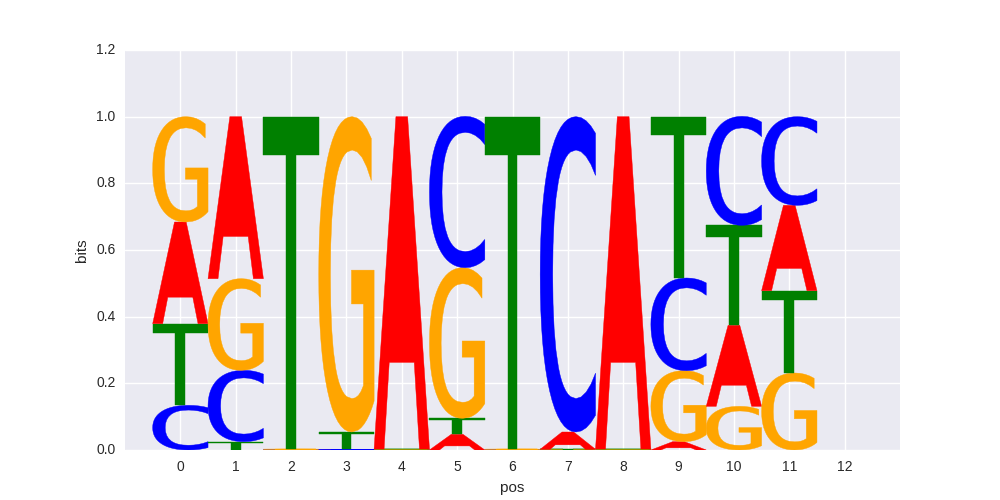 |
0.657489012293 |
>NFE2L2_2Nrf-2_transfac_M00821
|
 |
0.716082122463 |
FOSL2 |
 |
| 0.625972834049 |
NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer |
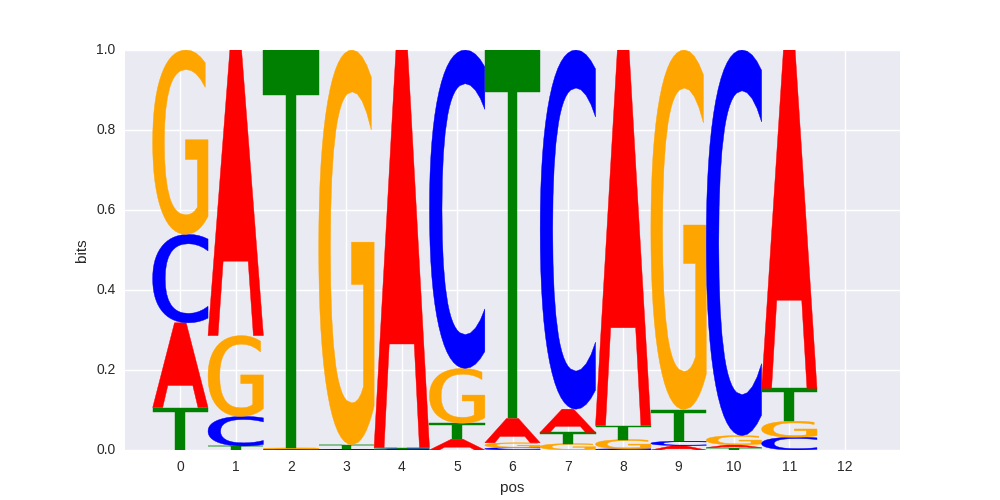 |
0.656606356901 |
>ATF3_known5CREB1_4Tax/CREB_transfac_M00115
|
 |
0.714603331126 |
JUND |
 |
| 0.622924099696 |
PAX3:FKHR-fusion(Paired,Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer |
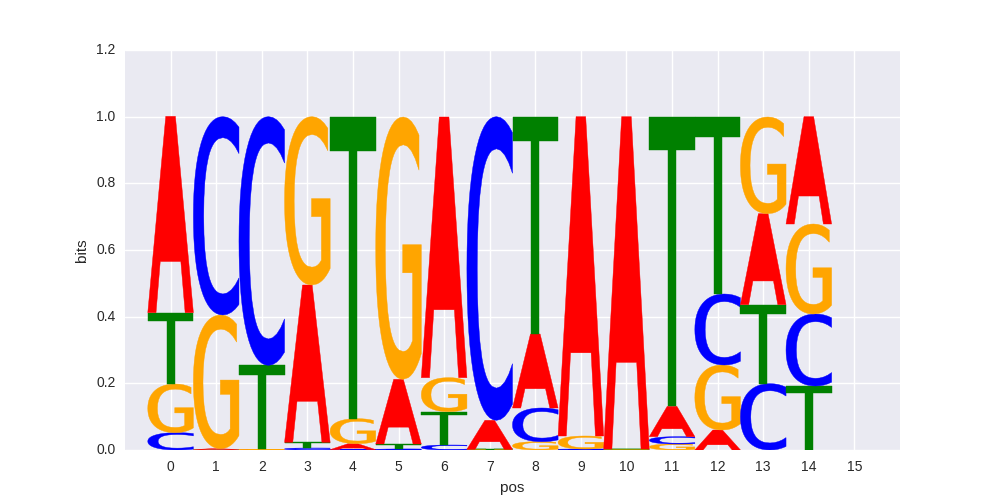 |
0.646398620665 |
>MAFG_1MAFG_jolma_full_M383
|
 |
0.712808422936 |
JUNB |
 |
| 0.62100248293 |
Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer |
 |
0.632607460036 |
>NFE2L1::MAFG_1TCF11:MafG_transfac_M00284
|
 |
0.712023058125 |
M2428_1.02.txt |
 |
| 0.619363485856 |
NFAT:AP1(RHD,bZIP)/Jurkat-NFATC1-ChIP-Seq(Jolma_et_al.)/Homer |
 |
0.627249319557 |
>SOX9_9SOX9_jolma_full_M796
|
 |
0.711853230517 |
FOS |
 |
| 0.616232883172 |
PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer |
 |
0.625745318225 |
>AP1_known7AP1_8AP-1_transfac_M00924
|
 |
0.711746311941 |
JUN |
 |