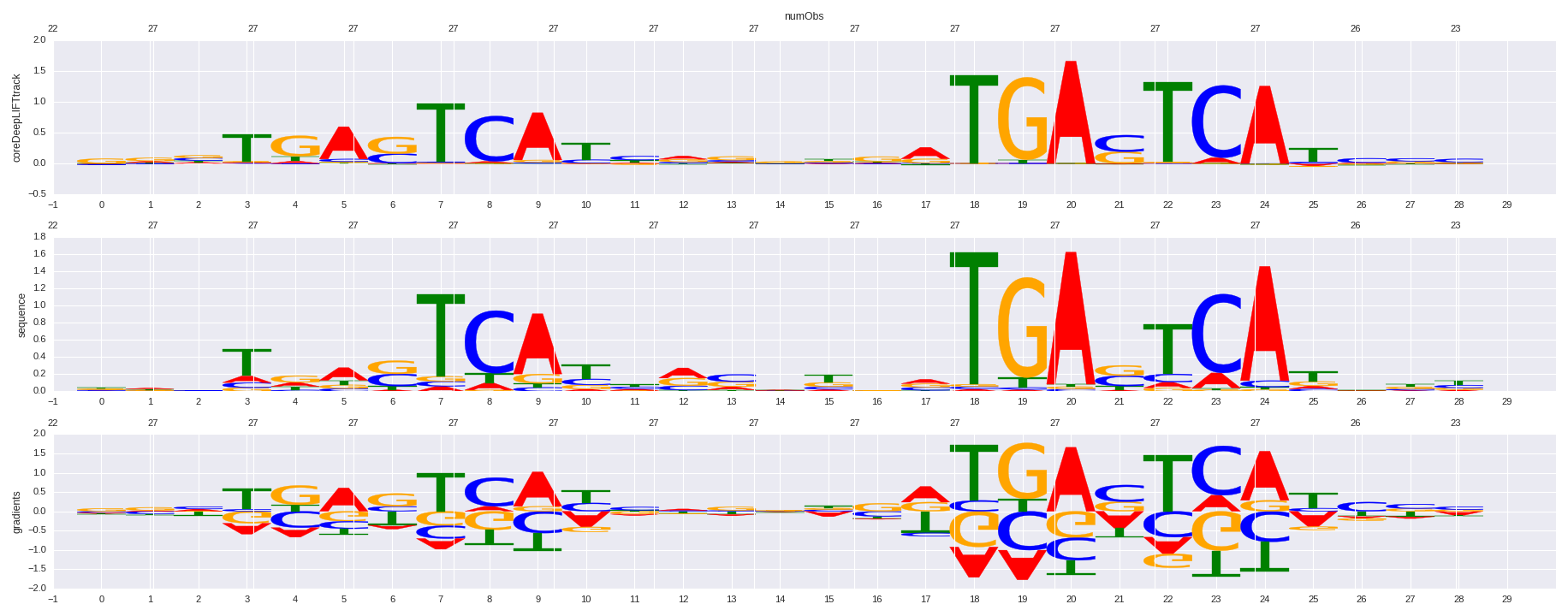
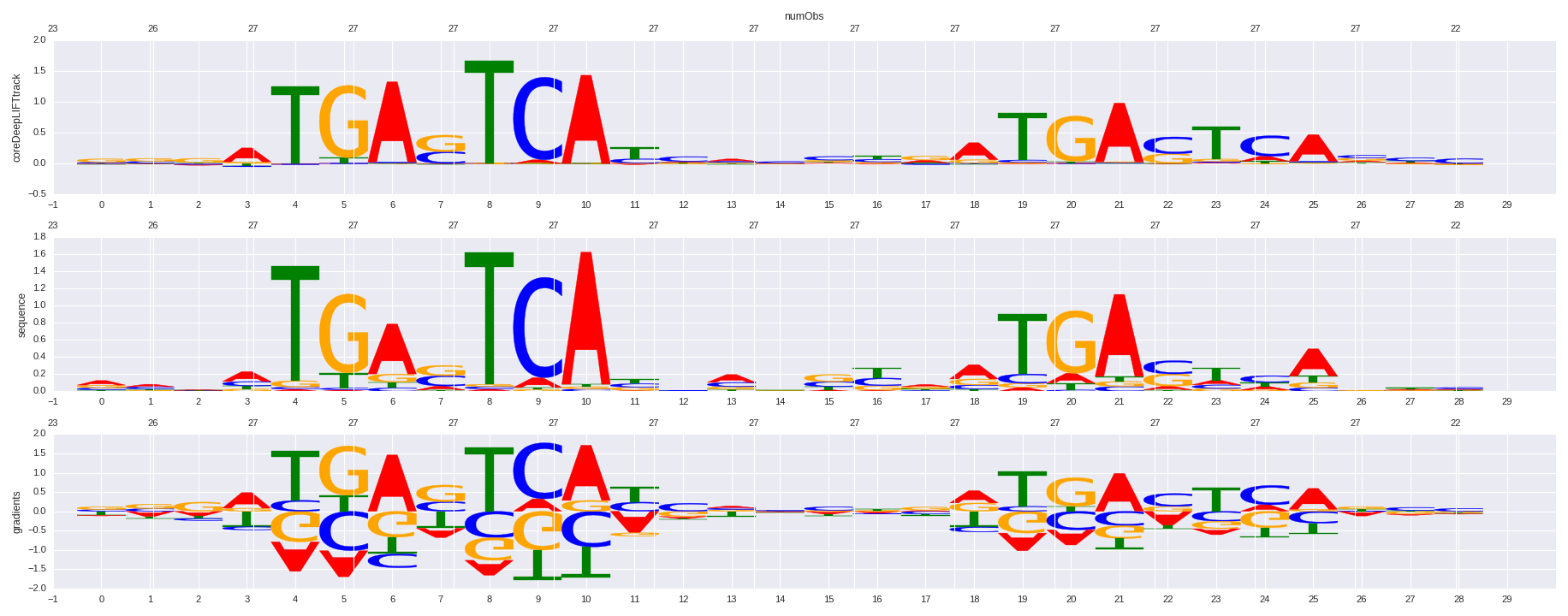
| HOMER correlation |
HOMER motif name |
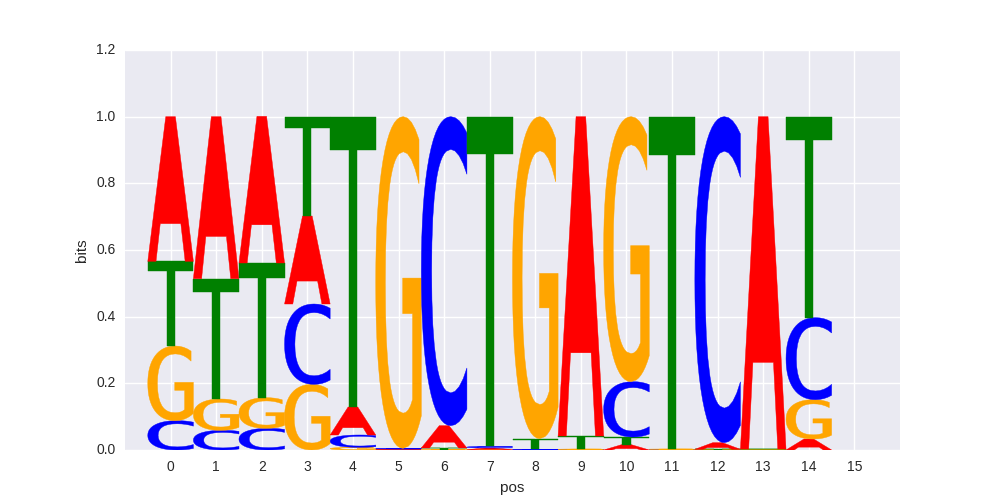
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
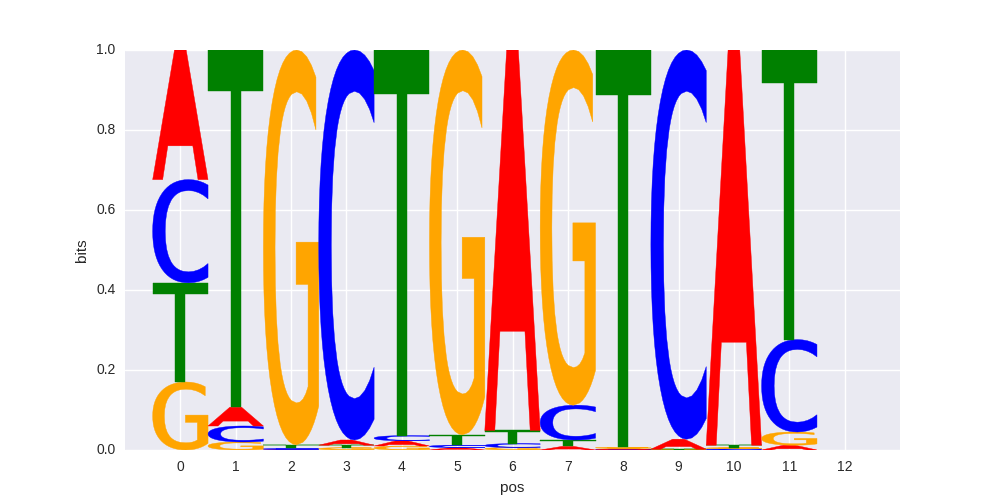
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.742402265887 |
Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer |
 |
0.768678072041 |
>BACH1_1Bach1_transfac_M00495
|
 |
0.768678074296 |
BACH1 |
 |
| 0.74176487963 |
Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer |
 |
0.746986542027 |
>BATF_disc1 BATF_GM12878_encode-Myers_seq_hsa_r1:AlignACE#1#Intergenic
|
 |
0.765239663584 |
JUND |
 |
| 0.653853995176 |
PAX3:FKHR-fusion(Paired,Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer |
 |
0.714136526114 |
>MAFF_1MAFF_jolma_DBD_M382
|
 |
0.762684229941 |
JUND |
 |
| 0.640186668045 |
PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer |
 |
0.685456016485 |
>NFE2L1::MAFG_1TCF11:MafG_transfac_M00284
|
 |
0.762472624402 |
M2428_1.02.txt |
 |
| 0.634071683437 |
OCT:OCT(POU,Homeobox,IR1)/NPC-Brn2-ChIP-Seq(GSE35496)/Homer |
 |
0.680219841287 |
>NFE2L2_2Nrf-2_transfac_M00821
|
 |
0.759790018614 |
FOS |
 |
| 0.627968894945 |
NFAT:AP1(RHD,bZIP)/Jurkat-NFATC1-ChIP-Seq(Jolma_et_al.)/Homer |
 |
0.678180372589 |
>MAFG_1MAFG_jolma_full_M383
|
 |
0.758514350101 |
FOSL2 |
 |
| 0.626672728749 |
ZFP3(Zf)/HEK293-ZFP3.GFP-ChIP-Seq(GSE58341)/Homer |
 |
0.675992838473 |
>ATF3_known5CREB1_4Tax/CREB_transfac_M00115
|
 |
0.757776829376 |
JUND |
 |
| 0.618992885678 |
Pit1+1bp(Homeobox)/GCrat-Pit1-ChIP-Seq(GSE58009)/Homer |
 |
0.675736823959 |
>VENTX_2VENTX_jolma_DBD_M650
|
 |
0.756631713606 |
FOS |
 |
| 0.614847211061 |
Pbx3(Homeobox)/GM12878-PBX3-ChIP-Seq(GSE32465)/Homer |
 |
0.667090072985 |
>SOX15_4SOX15_jolma_full_M765
|
 |
0.753774849756 |
JUNB |
 |
| 0.612724143614 |
Pknox1(Homeobox)/ES-Prep1-ChIP-Seq(GSE63282)/Homer |
 |
0.657567050044 |
>MAF_known11MAFB_4Mafb_jolma_DBD_M389
|
 |
0.753305305911 |
JUN |
 |