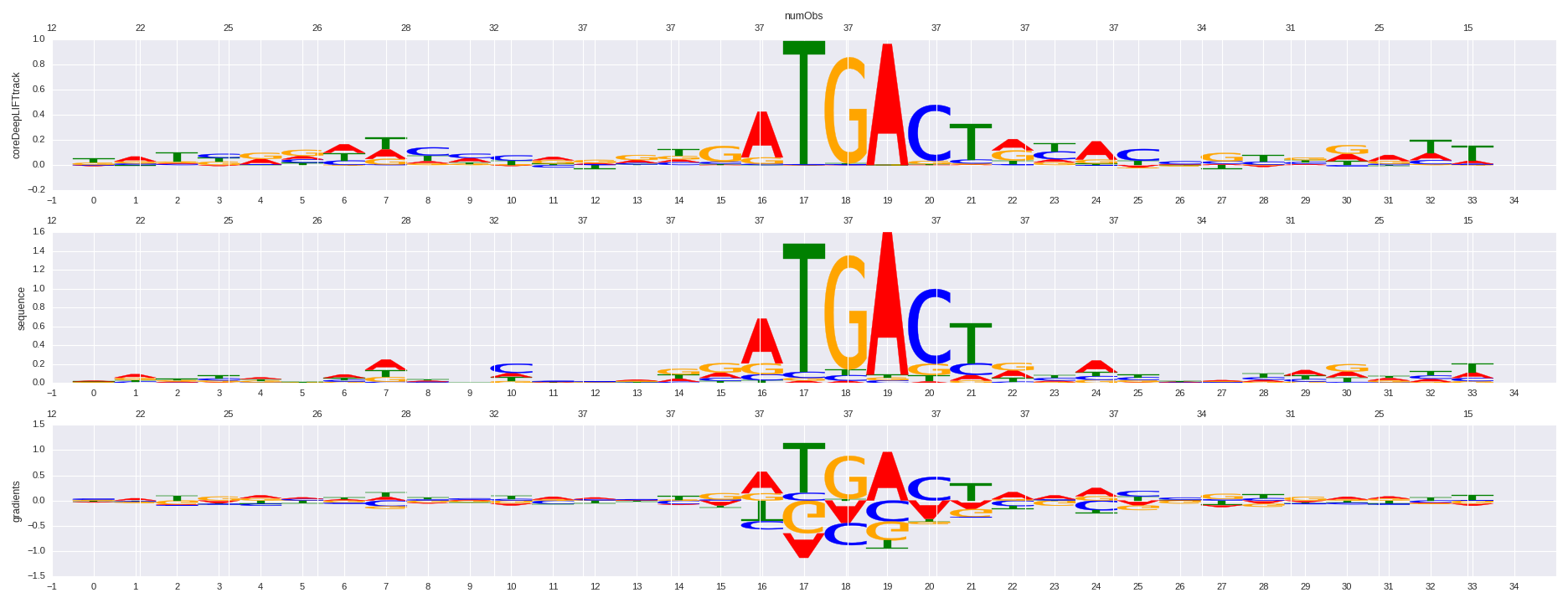
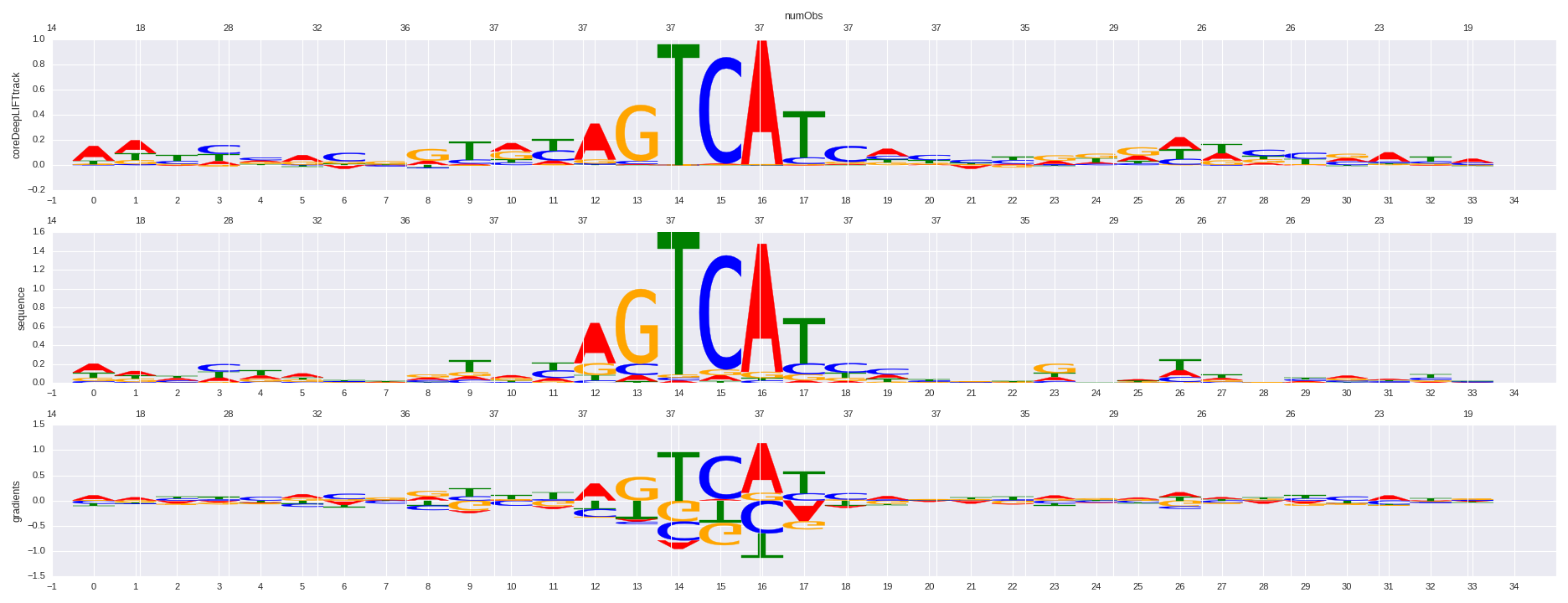
| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.802813321057 |
Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer |
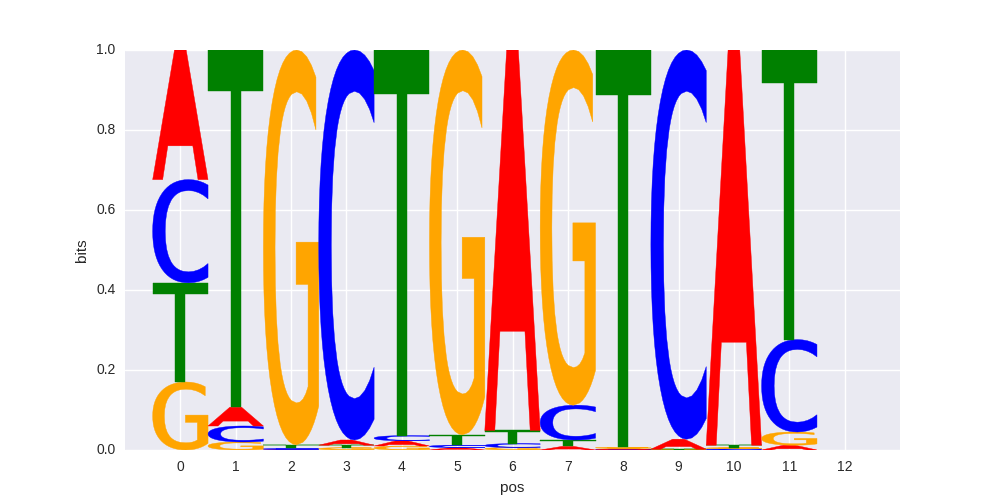 |
0.836304010191 |
>SOX9_6SOX9_jolma_full_M793
|
 |
0.836303992973 |
SOX9 |
 |
| 0.800915604033 |
NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer |
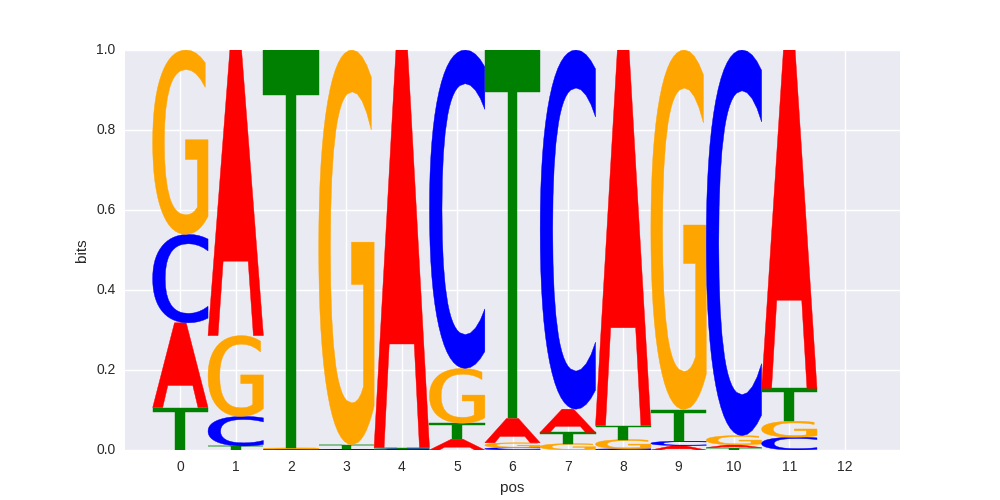 |
0.811503862192 |
>SOX10_6SOX10_jolma_full_M758
|
 |
0.811503842312 |
SOX10 |
 |
| 0.791424348678 |
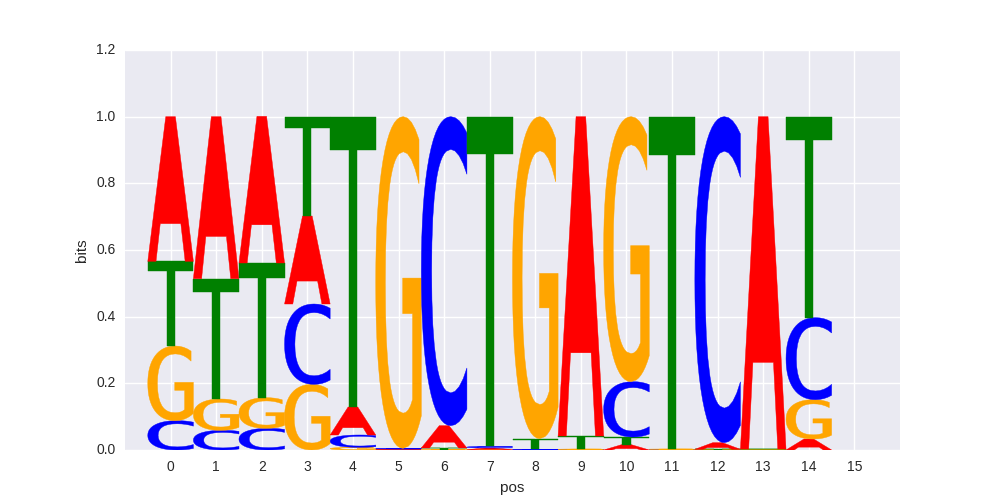
Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer |
 |
0.80898052934 |
>SOX10_10Sox10_jolma_DBD_M803
|
 |
0.80898051941 |
M6085_1.02.txt |
 |
| 0.768704536143 |
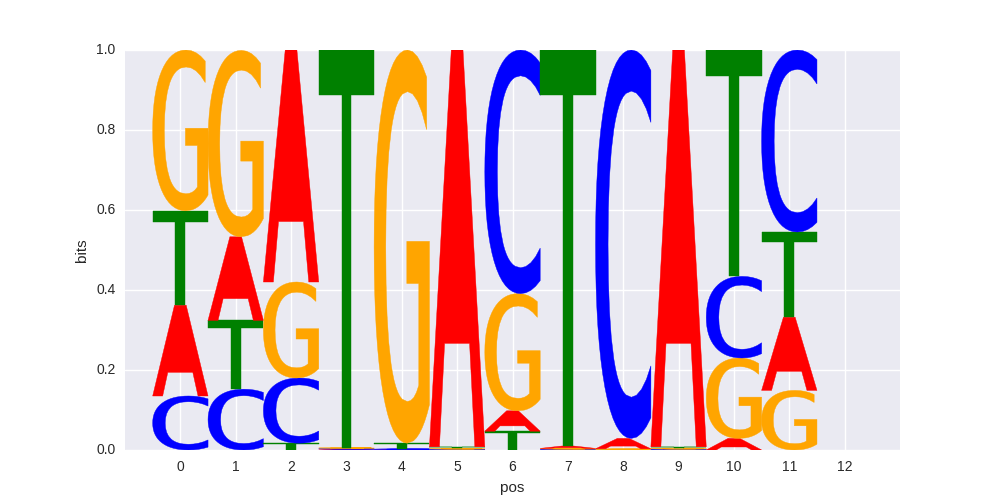
Fra2(bZIP)/Striatum-Fra2-ChIP-Seq(GSE43429)/Homer |
 |
0.803731642301 |
>SOX10_7SOX10_jolma_full_M759
|
 |
0.803731632188 |
SOX10 |
 |
| 0.761736860209 |
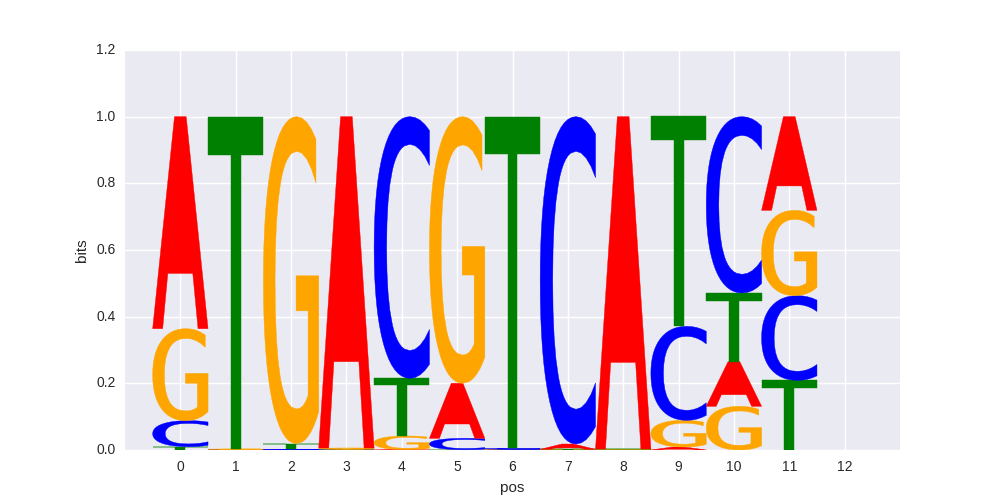
JunD(bZIP)/K562-JunD-ChIP-Seq/Homer |
 |
0.801665004946 |
>SOX9_9SOX9_jolma_full_M796
|
 |
0.801664992792 |
SOX9 |
 |
| 0.754075188922 |
Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer |
 |
0.800303446721 |
>SOX8_6SOX8_jolma_DBD_M787
|
 |
0.800303390019 |
SOX8 |
 |
| 0.753440966682 |
AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer |
 |
0.798157455186 |
>BATF_known1BATF3_1BATF3_jolma_DBD_M355
|
 |
0.799521319503 |
NFE2 |
 |
| 0.751002816067 |
Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer |
 |
0.79306501296 |
>BACH1_1Bach1_transfac_M00495
|
 |
0.798157391329 |
BATF3 |
 |
| 0.749805529811 |
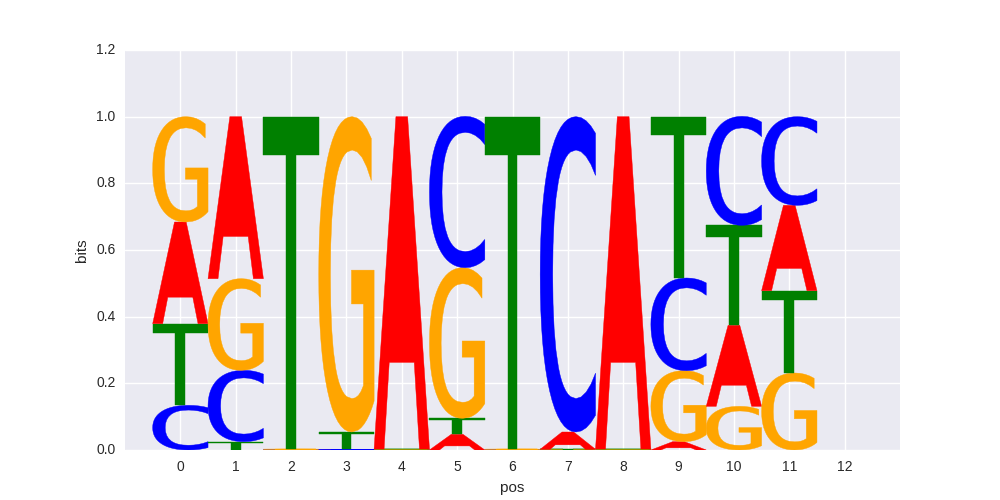
Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer |
 |
0.791784254891 |
>NFE2_known2NFE2_2NFE2_jolma_DBD_M391
|
 |
0.793065001541 |
BACH1 |
 |
| 0.749060283969 |
c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer |
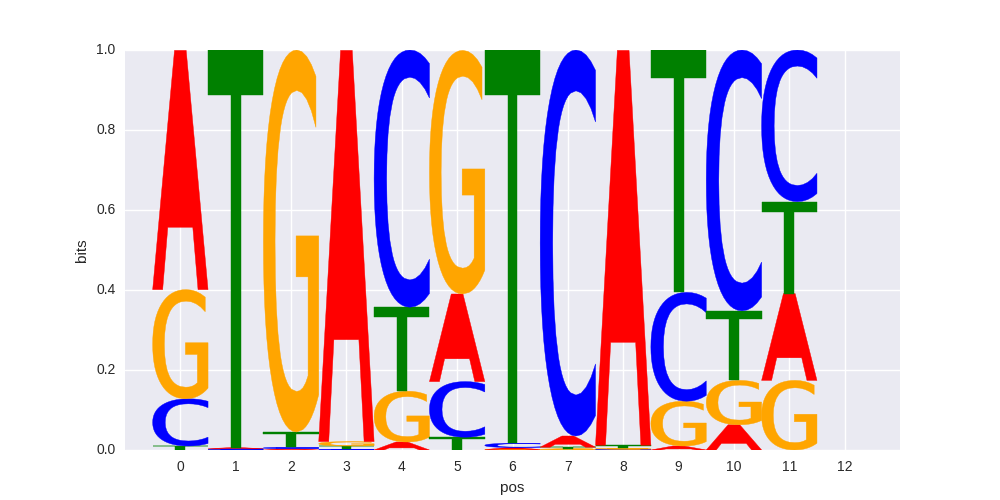 |
0.78881125261 |
>NFE2_disc1 NFE2_K562_encode-Snyder_seq_hsa_r1:AlignACE#1#Intergenic
|
 |
0.791784276206 |
NFE2 |
 |