| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
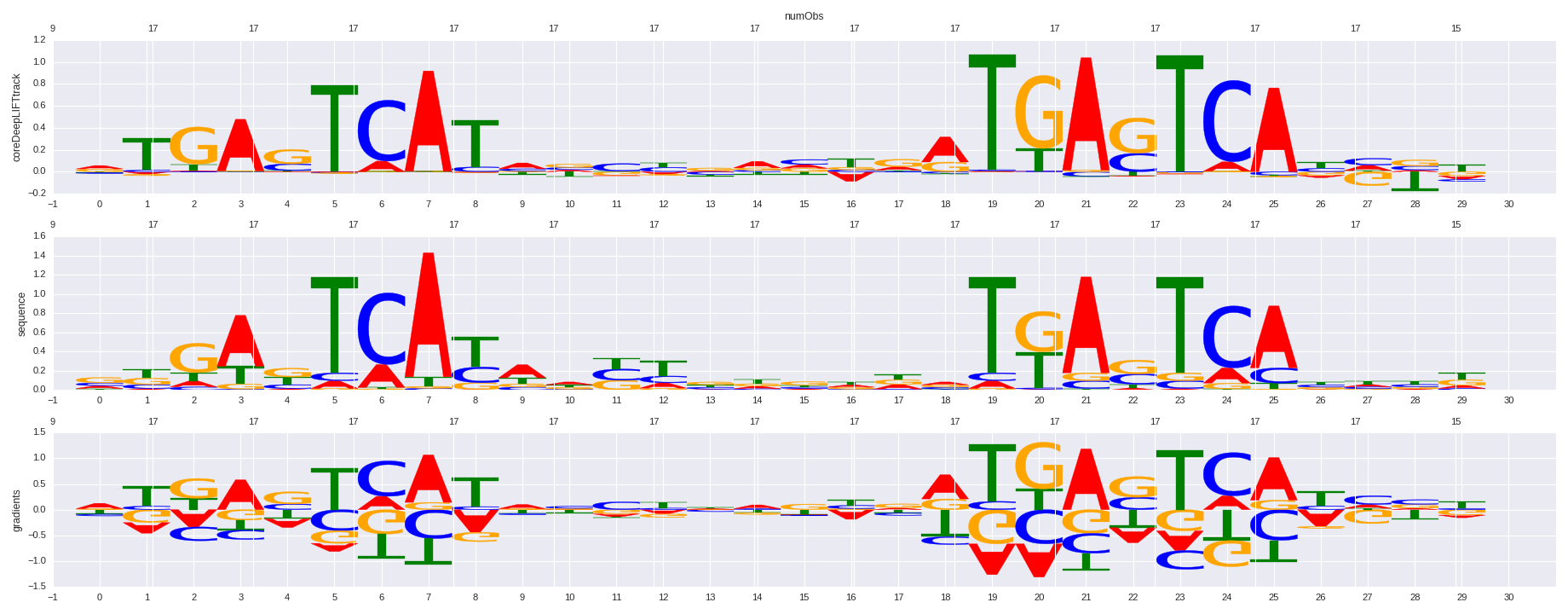
| 0.699440524729 |
Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer |
 |
0.722094502558 |
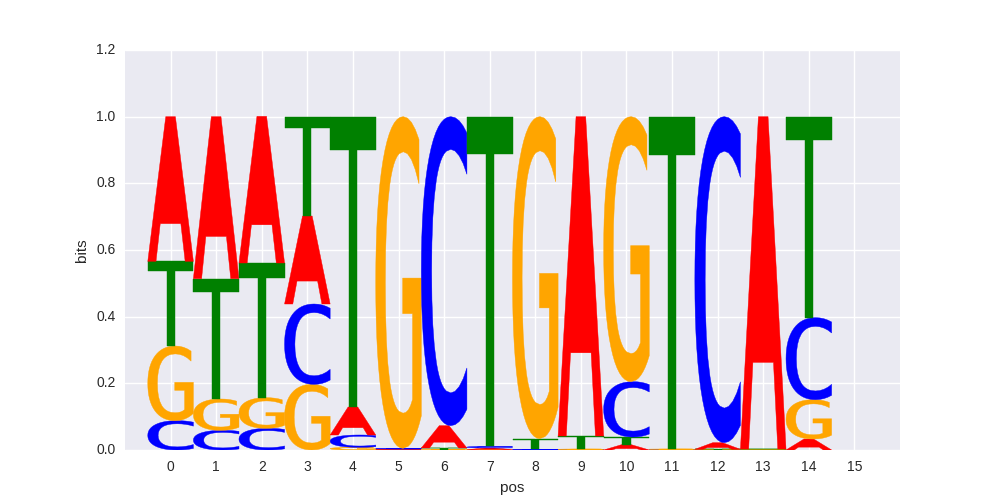
>BACH1_1Bach1_transfac_M00495
|
 |
0.727970027544 |
BACH1 |
 |
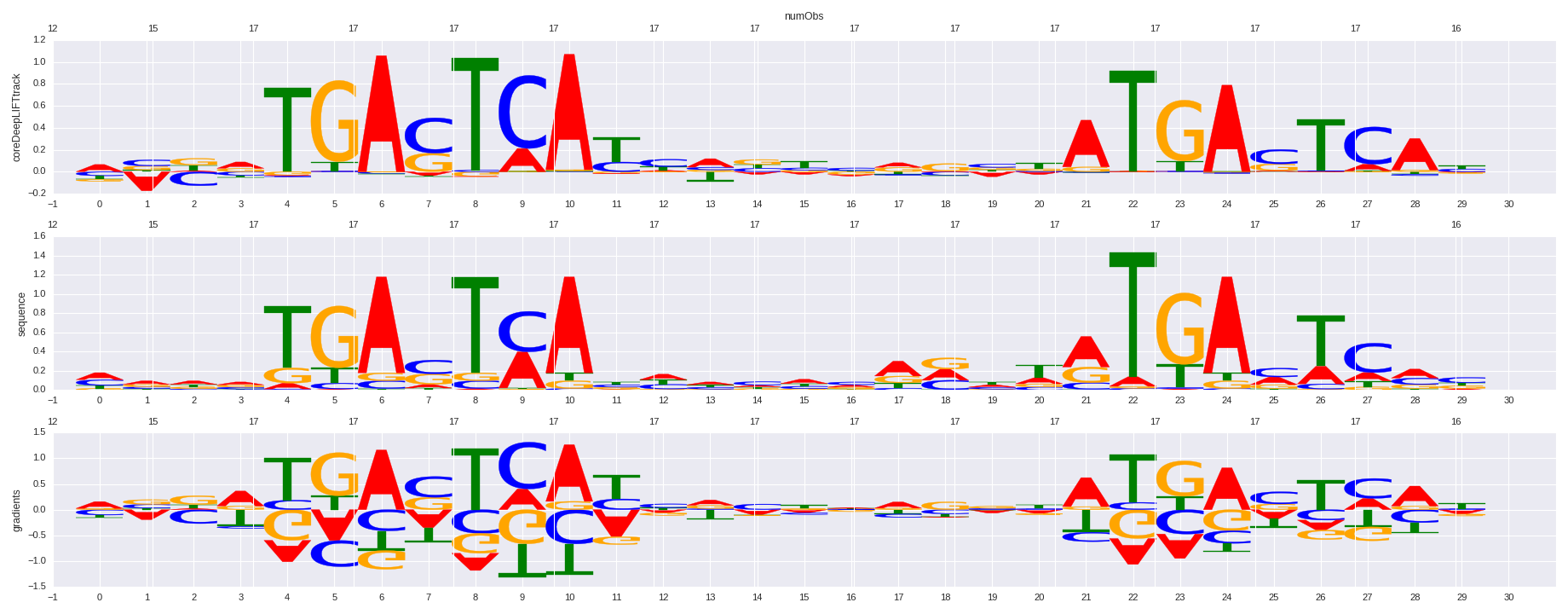
| 0.633853871292 |
PAX3:FKHR-fusion(Paired,Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer |
 |
0.671112172752 |
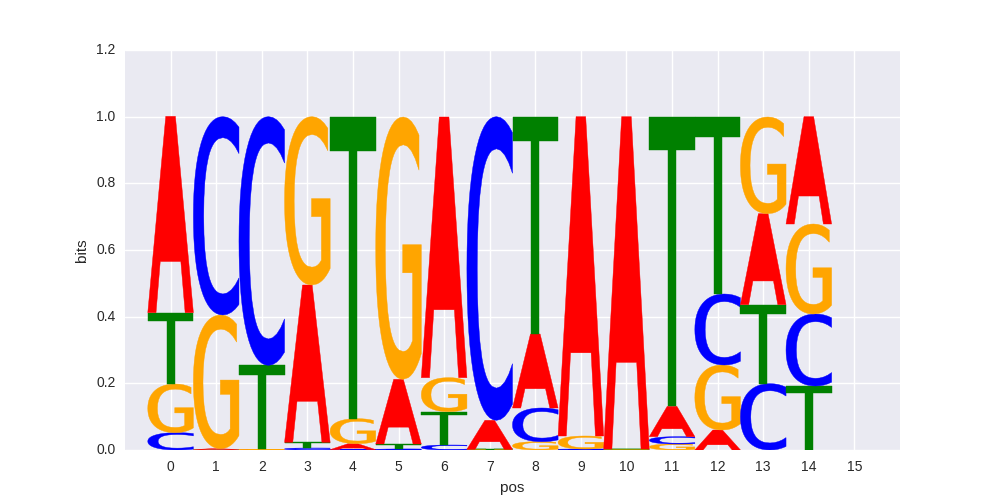
>VENTX_2VENTX_jolma_DBD_M650
|
 |
0.726495577269 |
JUND |
 |
| 0.616014359073 |
PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer |
 |
0.670255391683 |
>MAFF_1MAFF_jolma_DBD_M382
|
 |
0.723479294801 |
JUND |
 |
| 0.609024228015 |
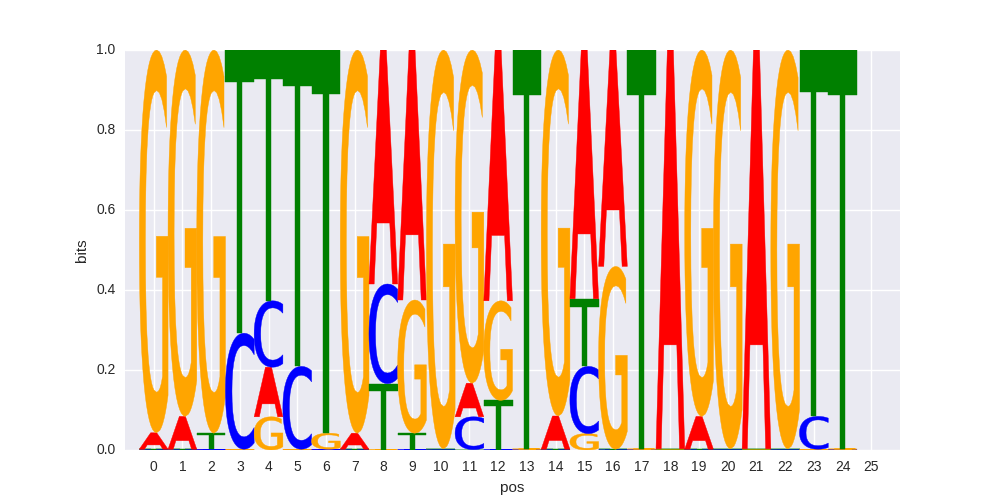
ZFP3(Zf)/HEK293-ZFP3.GFP-ChIP-Seq(GSE58341)/Homer |
 |
0.652951227511 |
>ATF3_known5CREB1_4Tax/CREB_transfac_M00115
|
 |
0.722370501968 |
FOS |
 |
| 0.605956975292 |
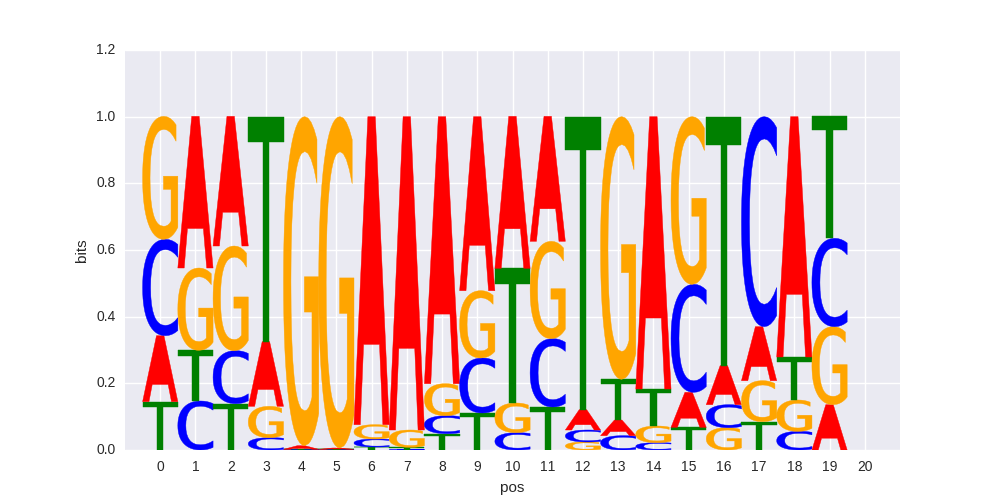
NFAT:AP1(RHD,bZIP)/Jurkat-NFATC1-ChIP-Seq(Jolma_et_al.)/Homer |
 |
0.651348431602 |
>NFE2L1::MAFG_1TCF11:MafG_transfac_M00284
|
 |
0.718364513909 |
JUNB |
 |
| 0.605045821535 |
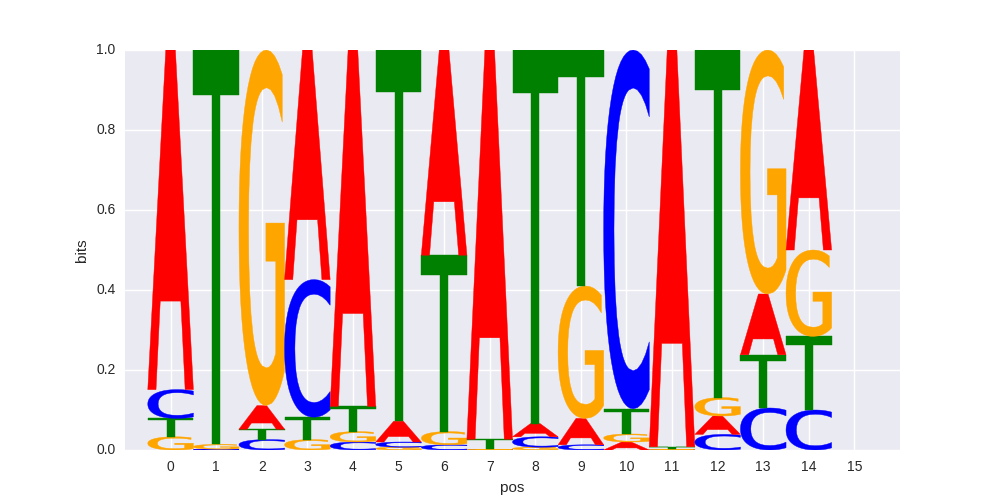
OCT:OCT(POU,Homeobox,IR1)/NPC-Brn2-ChIP-Seq(GSE35496)/Homer |
 |
0.643856390772 |
>MAFG_1MAFG_jolma_full_M383
|
 |
0.717587735926 |
FOSL2 |
 |
| 0.593464786083 |
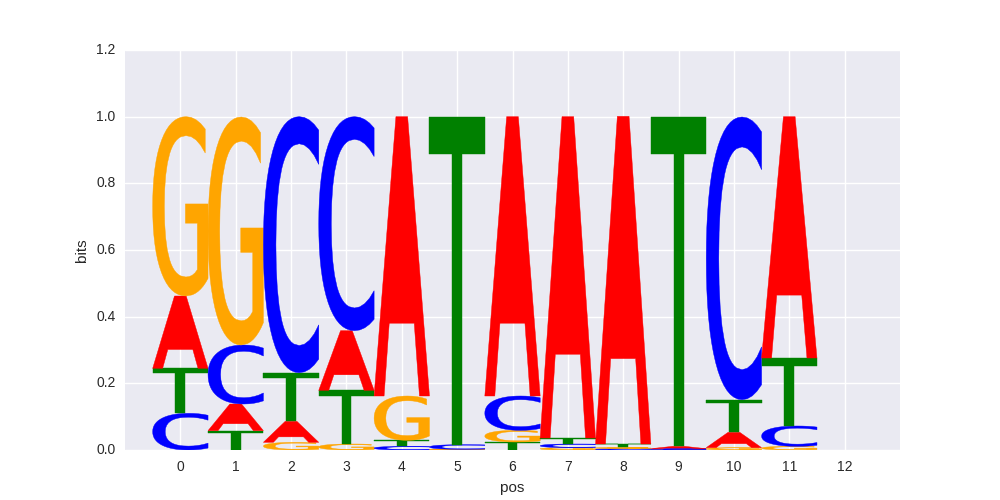
Hoxc9(Homeobox)/Ainv15-Hoxc9-ChIP-Seq(GSE21812)/Homer |
 |
0.634593173678 |
>AP1_disc5 FOS_K562_encode-White_seq_hsa_eGFP_r1:Trawler#3#Intergenic
|
 |
0.716986920267 |
M2428_1.02.txt |
 |
| 0.587220612647 |
Pit1+1bp(Homeobox)/GCrat-Pit1-ChIP-Seq(GSE58009)/Homer |
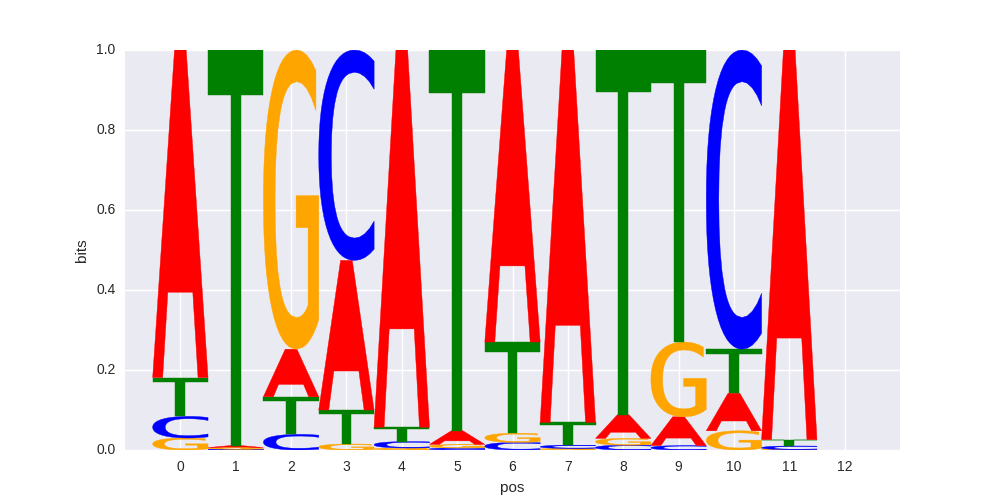 |
0.626414271149 |
>SOX15_4SOX15_jolma_full_M765
|
 |
0.716081414565 |
JUND |
 |
| 0.586861658367 |
Hoxb4(Homeobox)/ES-Hoxb4-ChIP-Seq(GSE34014)/Homer |
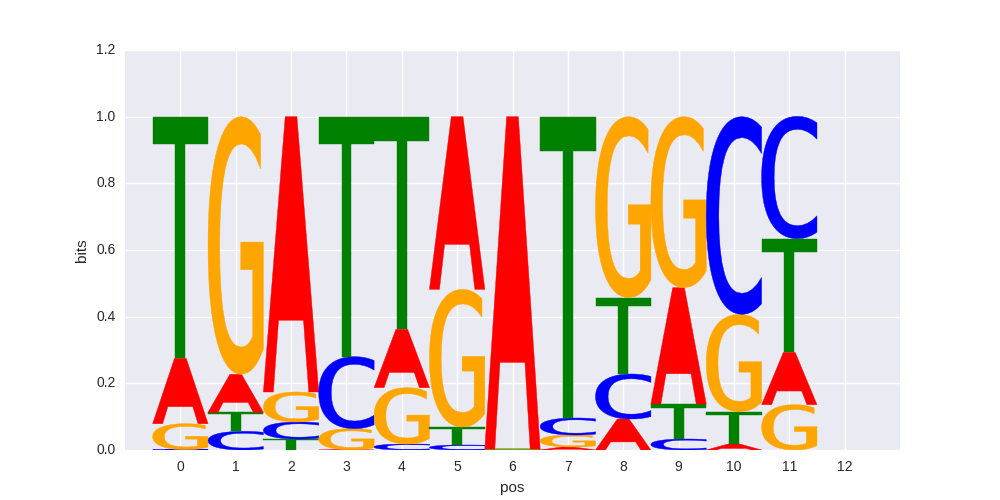 |
0.62483164948 |
>MAF_known11MAFB_4Mafb_jolma_DBD_M389
|
 |
0.712722114114 |
FOS |
 |
| 0.58470895783 |
OCT:OCT(POU,Homeobox)/NPC-Brn1-ChIP-Seq(GSE35496)/Homer |
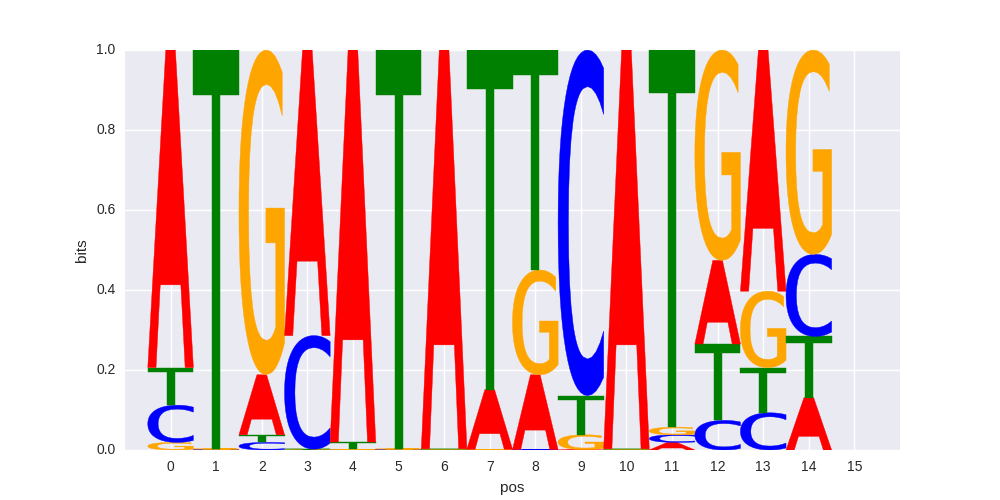 |
0.621704387806 |
>MAF_known9MAFK_5MAFK_jolma_full_M387
|
 |
0.711240094676 |
JUN |
 |