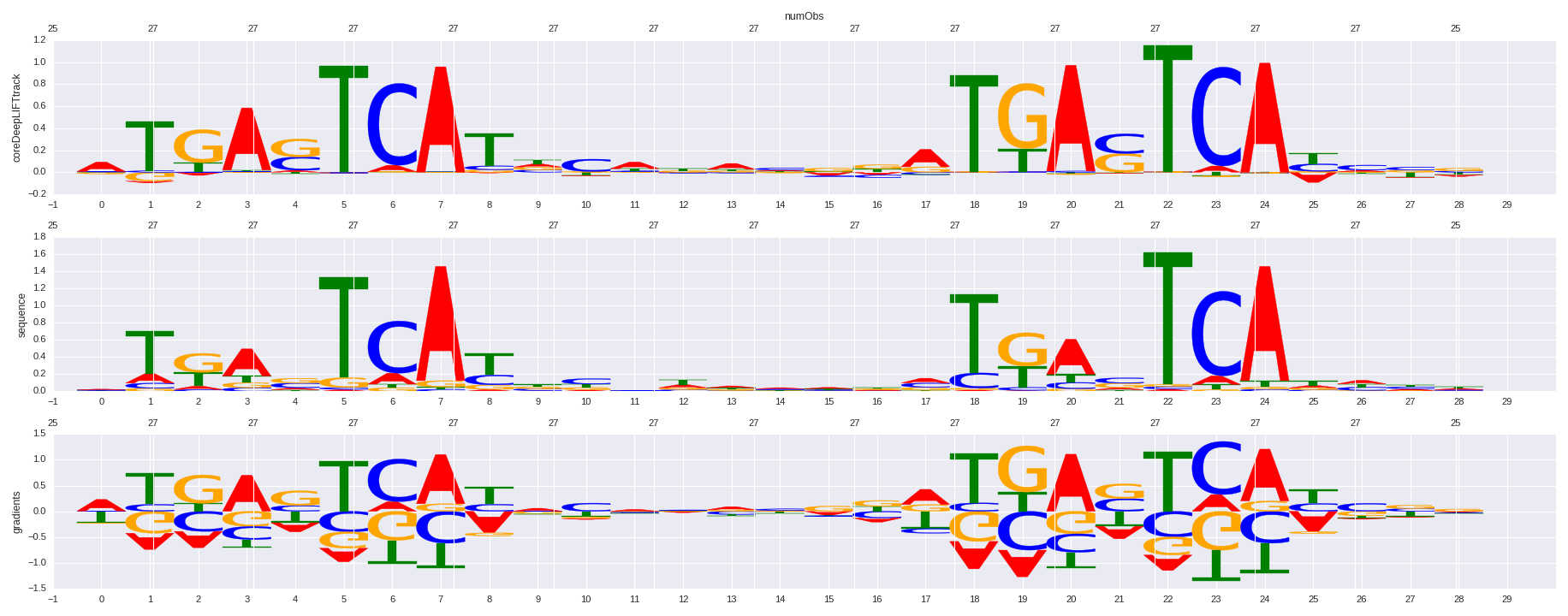
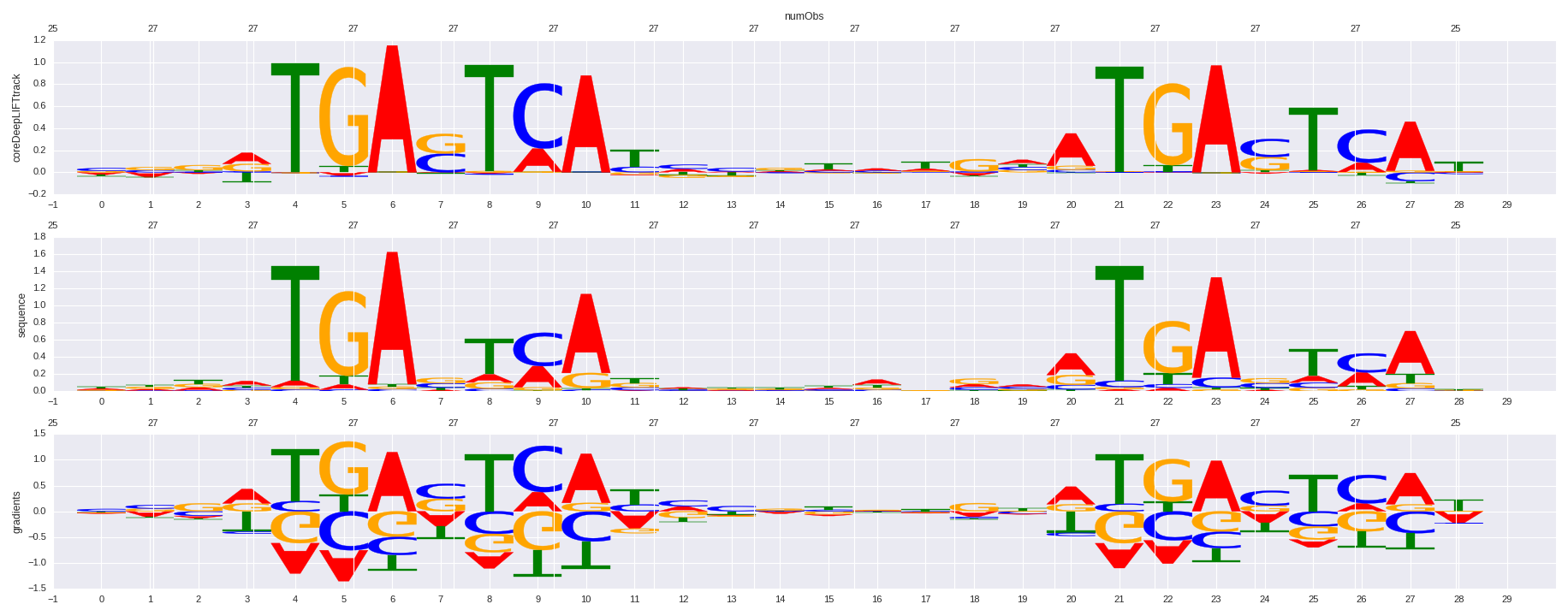
| HOMER correlation |
HOMER motif name |
HOMER motif PWM |
ENCODE correlation |
ENCODE motif name |
ENCODE motif PWM |
CISBP correlation |
CISBP motif name |
CISBP motif PWM |
| 0.694475306603 |
Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer |
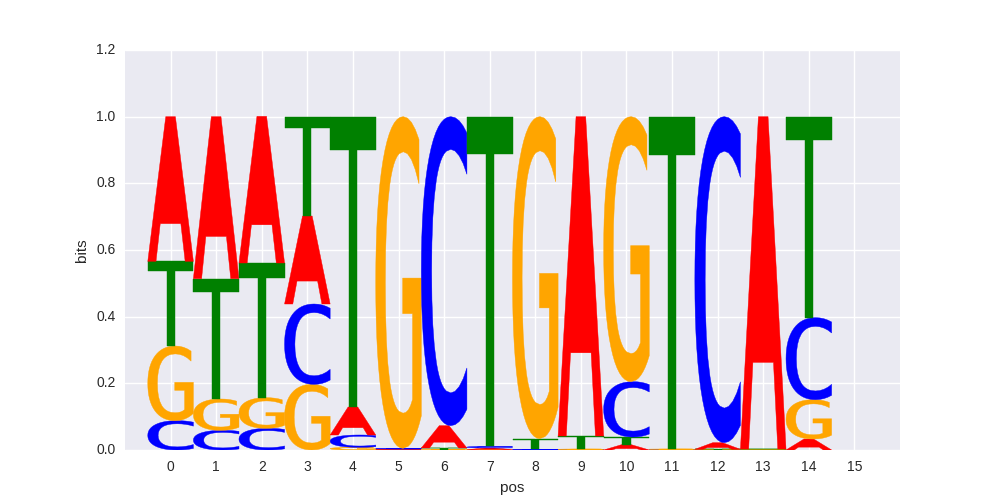 |
0.712321939821 |
>BACH1_1Bach1_transfac_M00495
|
 |
0.715452012669 |
JUND |
 |
| 0.692119393745 |
Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer |
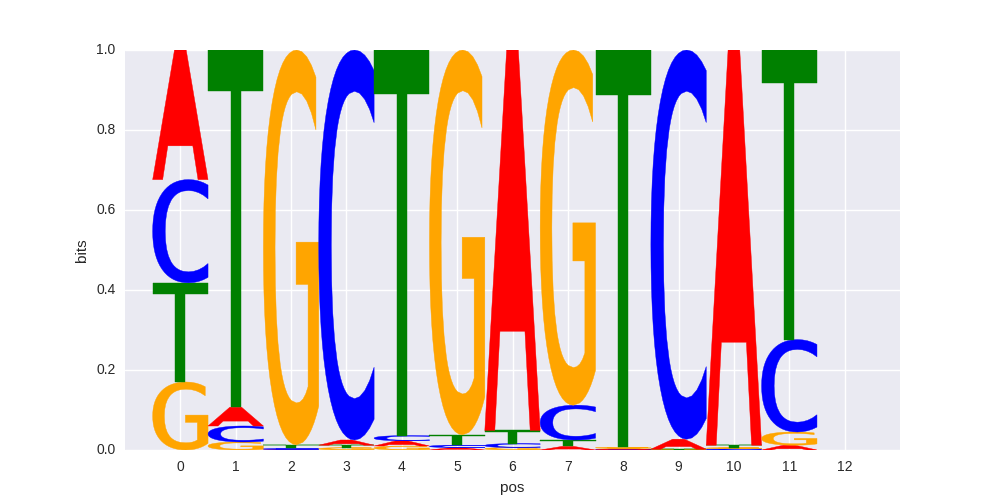 |
0.697447517566 |
>BATF_disc1 BATF_GM12878_encode-Myers_seq_hsa_r1:AlignACE#1#Intergenic
|
 |
0.713419380162 |
JUND |
 |
| 0.619359386256 |
PAX3:FKHR-fusion(Paired,Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer |
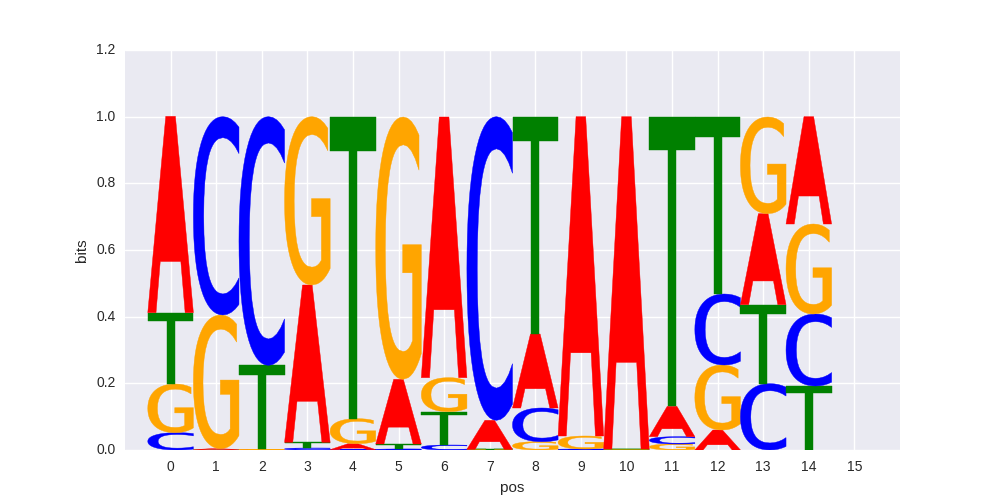 |
0.66498372471 |
>MAFF_1MAFF_jolma_DBD_M382
|
 |
0.712321940072 |
BACH1 |
 |
| 0.61786663847 |
NFAT:AP1(RHD,bZIP)/Jurkat-NFATC1-ChIP-Seq(Jolma_et_al.)/Homer |
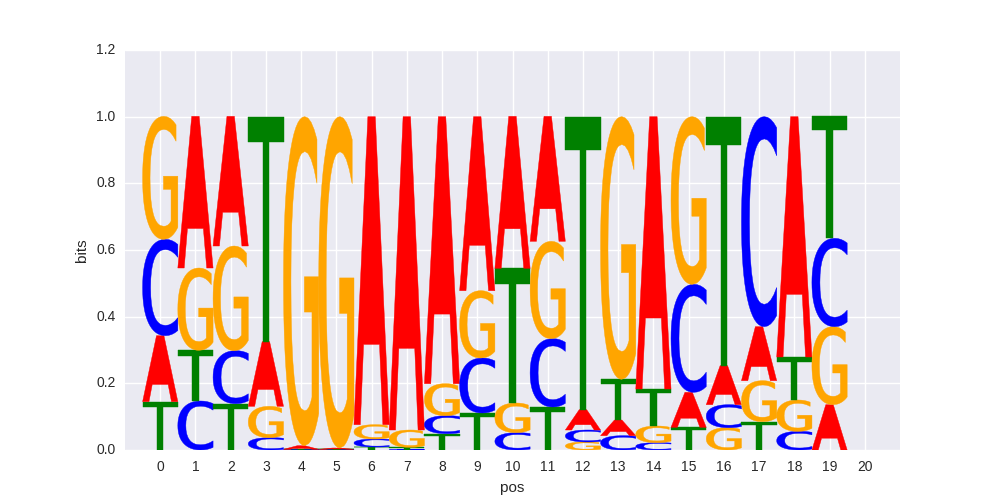 |
0.662520434188 |
>AIRE_2AIRE_transfac_M01000
|
 |
0.711679909722 |
FOS |
 |
| 0.612514322606 |
PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer |
 |
0.654090674415 |
>ATF3_known5CREB1_4Tax/CREB_transfac_M00115
|
 |
0.708869837907 |
FOSL2 |
 |
| 0.604892032955 |
ZFP3(Zf)/HEK293-ZFP3.GFP-ChIP-Seq(GSE58341)/Homer |
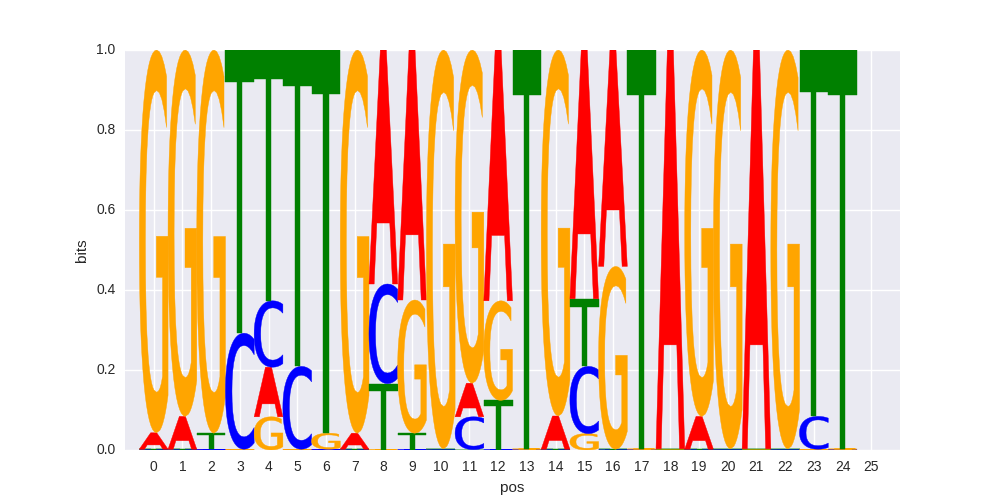 |
0.646080026011 |
>SOX15_4SOX15_jolma_full_M765
|
 |
0.707921962721 |
JUNB |
 |
| 0.602762907322 |
OCT:OCT(POU,Homeobox,IR1)/NPC-Brn2-ChIP-Seq(GSE35496)/Homer |
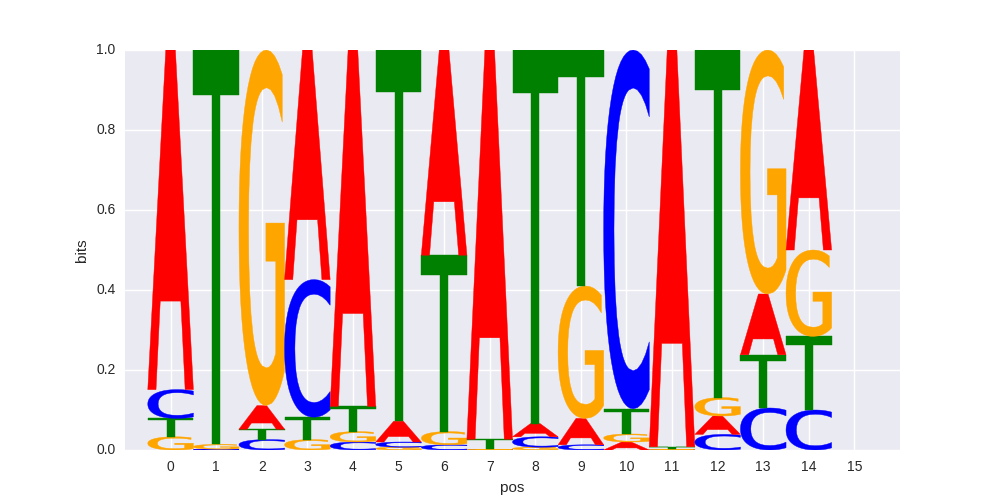 |
0.643803069881 |
>NFE2L2_2Nrf-2_transfac_M00821
|
 |
0.707570098853 |
JUND |
 |
| 0.591678084791 |
Pbx3(Homeobox)/GM12878-PBX3-ChIP-Seq(GSE32465)/Homer |
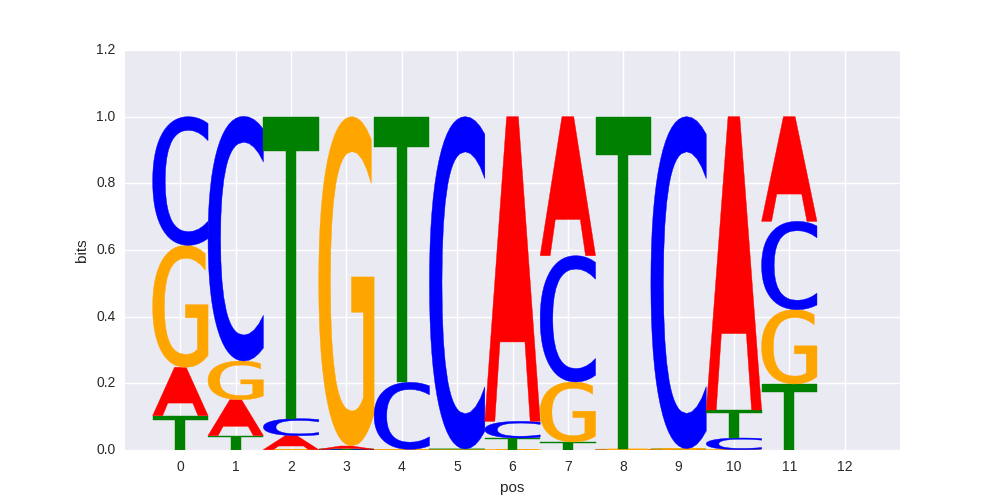 |
0.636947126338 |
>NFE2L1::MAFG_1TCF11:MafG_transfac_M00284
|
 |
0.707140290045 |
M2428_1.02.txt |
 |
| 0.59080118131 |
Pknox1(Homeobox)/ES-Prep1-ChIP-Seq(GSE63282)/Homer |
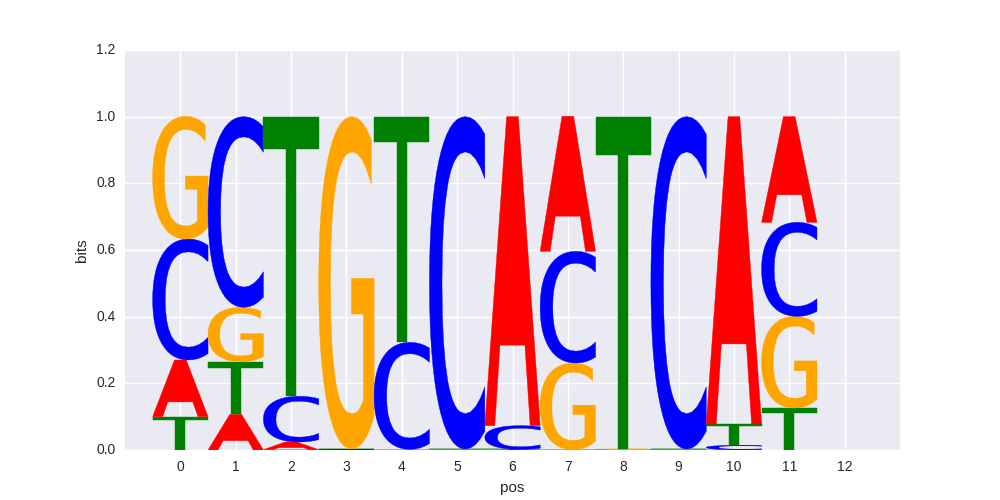 |
0.636477756696 |
>MAFG_1MAFG_jolma_full_M383
|
 |
0.706394395253 |
FOS |
 |
| 0.584423126885 |
Pit1+1bp(Homeobox)/GCrat-Pit1-ChIP-Seq(GSE58009)/Homer |
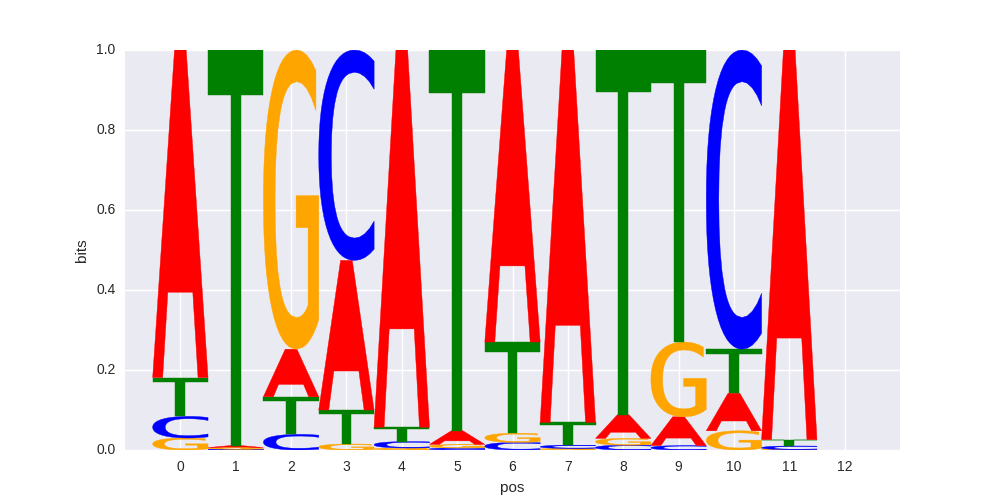 |
0.62543892863 |
>T_1Brachyury_transfac_M00150
|
 |
0.703389834544 |
JUN |
 |